

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.4 - phase: 0
(581 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
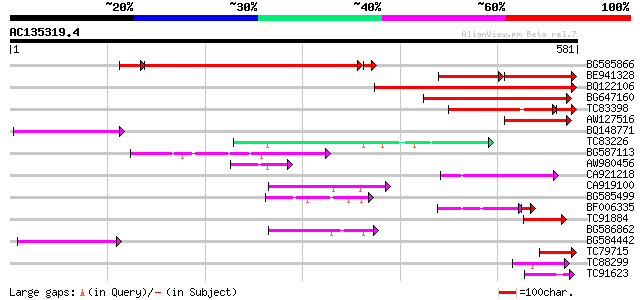
Score E
Sequences producing significant alignments: (bits) Value
BG585866 286 6e-86
BE941328 114 2e-52
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 179 2e-45
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 175 5e-44
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 117 4e-29
AW127516 82 6e-16
BQ148771 77 1e-14
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 76 4e-14
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 64 2e-10
AW980456 52 8e-07
CA921218 51 1e-06
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 50 2e-06
BG585499 49 5e-06
BF006335 similar to GP|14334538|gb| unknown protein {Arabidopsis... 44 3e-05
TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Bras... 46 3e-05
BG586862 45 1e-04
BG584442 44 1e-04
TC79715 similar to PIR|C86277|C86277 hypothetical protein AAF439... 43 4e-04
TC88299 weakly similar to GP|17979137|gb|AAL49826.1 unknown prot... 43 4e-04
TC91623 similar to PIR|D86241|D86241 protein T16B5.8 [imported] ... 42 7e-04
>BG585866
Length = 828
Score = 286 bits (731), Expect(3) = 6e-86
Identities = 128/223 (57%), Positives = 162/223 (72%)
Frame = +3
Query: 139 IIRAKNILKDGYTWRIGSGSSSFWFSNWNSLSPIGNYVPVIDVHDLQLSVRDVLSAPGNH 198
IIR KN+LK GYTWR GSG+SSFW++NW+SL +G P +D+HDL L+V+DV + G H
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNSSFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQH 263
Query: 199 ARVLYTNLPPDVENHINNIHTQFNDNIEDALIWSQNKNGTYTTKSGFNWLITSRVPAIEP 258
+ LYT LP D+ INN H FN +I DA IW N NG YT KSG++W++ S+ +
Sbjct: 264 TQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWIL-SQTETVNY 440
Query: 259 ILHAWSWIWRLKVPEKYKFLIWLACQNAVPTLSLLYHRSMAPSPICTRCGEEDETFMHCV 318
+WSWIWRLK+PEKYKF +WLAC NAVPTLSLL HR+M S IC+RCGE +E+F HCV
Sbjct: 441 NNSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCV 620
Query: 319 RDCCFSKDIWQKIGFTSNDFVTTATTYDWIKLGISGSRSSIFL 361
RDC FSK IW KIGF+S DF ++++ DW+K GIS R + FL
Sbjct: 621 RDCRFSKIIWHKIGFSSPDFFSSSSVQDWLKDGISCHRPTTFL 749
Score = 37.7 bits (86), Expect(3) = 6e-86
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = +2
Query: 363 GLWWVWRHRNLMCL 376
GLWW+WRHR LMCL
Sbjct: 752 GLWWIWRHRTLMCL 793
Score = 34.3 bits (77), Expect(3) = 6e-86
Identities = 16/28 (57%), Positives = 21/28 (74%), Gaps = 1/28 (3%)
Frame = +1
Query: 113 LSNKYTGGHNTLY-ASANNRSSPTWSSI 139
L N Y+ GHN L+ A+A+ SSP+WSSI
Sbjct: 1 LFNNYSSGHNFLHIAAAHRNSSPSWSSI 84
>BE941328
Length = 505
Score = 114 bits (285), Expect(2) = 2e-52
Identities = 56/73 (76%), Positives = 59/73 (80%)
Frame = +2
Query: 508 RYHIYAALIQNIKDLLNDRNISLHHTLREGNQCADFFAKFGANSDAALVVHQSPPADLLP 567
RYH Y LIQNIKDLL RNISLHHTLR NQCADFFAK G NSDA LVVHQSPPADLLP
Sbjct: 206 RYHTYVVLIQNIKDLLIVRNISLHHTLR*RNQCADFFAKLGVNSDAHLVVHQSPPADLLP 385
Query: 568 LLRADALGIYFVR 580
LLR DA+ F++
Sbjct: 386 LLRTDAIDTVFIK 424
Score = 110 bits (274), Expect(2) = 2e-52
Identities = 52/67 (77%), Positives = 59/67 (87%)
Frame = +3
Query: 440 GYGGVLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSDSLLAV 499
GYGG+LRNSA LFIS FSG IPNS DIL A+LTAIHQGL ++ DLN+N++MCYSDSLLAV
Sbjct: 3 GYGGILRNSASLFISSFSGFIPNSTDIL*AKLTAIHQGLNMIIDLNMNDVMCYSDSLLAV 182
Query: 500 NLIANDT 506
NLI NDT
Sbjct: 183 NLIMNDT 203
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 179 bits (455), Expect = 2e-45
Identities = 94/206 (45%), Positives = 129/206 (61%)
Frame = -2
Query: 375 CLNNETLSLFRLCNNIISAADAANSAFNGEENPPHSDRLVKWNNRNHQDFILNVDGSCIG 434
CLNNE L L + + F +D +VKWN N+ +ILNVDGSC+G
Sbjct: 692 CLNNEEWLLIDLHSIFKTLLMLTAQPFLYNSLSQATDTVVKWNCDNNSCYILNVDGSCLG 513
Query: 435 TPPRTGYGGVLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSD 494
P TG+ G++RN AGLF SGF G I N++DILLAEL AI QGL++++D+ +++ +CY D
Sbjct: 512 NP*PTGFNGLIRNIAGLFNSGFPGNITNTSDILLAELHAIFQGLRMISDMGISDFVCYFD 333
Query: 495 SLLAVNLIANDTPRYHIYAALIQNIKDLLNDRNISLHHTLREGNQCADFFAKFGANSDAA 554
SL V+LI + ++H+YA LIQ+IKDL+ S+ HTL EGN CADF GA SD+
Sbjct: 332 SLHYVSLINGPSMKFHVYATLIQDIKDLVITSKASVFHTLCEGNYCADFLEMLGAASDSV 153
Query: 555 LVVHQSPPADLLPLLRADALGIYFVR 580
L +H SPP ++ L++ A F R
Sbjct: 152 LTIHVSPPDGMIQLIKVGAWETLFSR 75
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 175 bits (443), Expect = 5e-44
Identities = 83/151 (54%), Positives = 106/151 (69%)
Frame = +1
Query: 425 ILNVDGSCIGTPPRTGYGGVLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDL 484
ILNV+ SCIG P T +GGV+RN +I+GFSG I +I+ AELT +HQ LKL L
Sbjct: 328 ILNVERSCIGVPIYT*FGGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISL 507
Query: 485 NVNEIMCYSDSLLAVNLIANDTPRYHIYAALIQNIKDLLNDRNISLHHTLREGNQCADFF 544
N+ E++CYSDSLL VNLI D ++H+YA LI NIK +++ RN +LHHTLREGNQCADF
Sbjct: 508 NIEEMVCYSDSLLTVNLIKEDISQHHVYAVLIHNIKYIMSSRNFTLHHTLREGNQCADFM 687
Query: 545 AKFGANSDAALVVHQSPPADLLPLLRADALG 575
K ++D L +H SPP LLP +R D +G
Sbjct: 688 VKLRTSTDVDLTIHSSPPKVLLPSVRTDEIG 780
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 117 bits (294), Expect(2) = 4e-29
Identities = 63/112 (56%), Positives = 78/112 (69%)
Frame = +2
Query: 450 GLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSDSLLAVNLIANDTPRY 509
G F SGFSG I +S DIL AEL AI GL+L LN+ +++CYSDSL VNLI + Y
Sbjct: 89 GFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPSVVY 268
Query: 510 HIYAALIQNIKDLLNDRNISLHHTLREGNQCADFFAKFGANSDAALVVHQSP 561
H YA LIQ+IKDL+ +S HTLREGN+CADF AK GA+ D+ L +H +P
Sbjct: 269 HAYATLIQDIKDLI---RLSKLHTLREGNRCADFLAKLGASVDSTLPIHATP 415
Score = 28.9 bits (63), Expect(2) = 4e-29
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +3
Query: 561 PPADLLPLLRADALGIYFVR 580
PP LLPLL+ DA G YF R
Sbjct: 411 PPYGLLPLLKDDAWGTYFPR 470
>AW127516
Length = 379
Score = 82.0 bits (201), Expect = 6e-16
Identities = 36/68 (52%), Positives = 50/68 (72%)
Frame = +3
Query: 508 RYHIYAALIQNIKDLLNDRNISLHHTLREGNQCADFFAKFGANSDAALVVHQSPPADLLP 567
++H YA +Q+IKD + R+ ++ HTLREGN CAD+ AK GA+S++ VH +PP DLL
Sbjct: 171 KFHAYAVTVQDIKDTIASRSFTIQHTLREGNHCADYMAKLGASSNSDFSVHLTPPHDLLG 350
Query: 568 LLRADALG 575
LLR DA+G
Sbjct: 351 LLRNDAIG 374
>BQ148771
Length = 680
Score = 77.4 bits (189), Expect = 1e-14
Identities = 40/114 (35%), Positives = 61/114 (53%), Gaps = 1/114 (0%)
Frame = -3
Query: 5 LASWKNRLLNKTGRLTLANSVLTSIPSYFMQTNWLPQNICDSIDQTTRNFIWRGT-TNKG 63
LA+WK L+ R+TLA SV+ ++P Y M T +P+ + I + R F+W T ++
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 64 IHLVNWKKISLPKNIGGLSIRAAREANTSLLGKLVWDMVQSSNKLWVSLLSNKY 117
H V W+ +S PK I GL +R N + + KL W + SN L ++ KY
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKY 232
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 75.9 bits (185), Expect = 4e-14
Identities = 69/292 (23%), Positives = 116/292 (39%), Gaps = 26/292 (8%)
Frame = +3
Query: 230 IWSQNKNGTYTTKSGFNWLITSRVPAIEPILHA------WSWIWRLKVPEKYKFLIWLAC 283
+W N G Y+ KSG+N L T + I + W IW L ++K L+W
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 284 QNAVPTLSLLYHRSMAPSPICTRCGEEDETFMHCVRDCCFSKDIWQKIGFTSN-DFVTTA 342
+++P S L R + P+C RC + ET H C SK +W N D +
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPNP 362
Query: 343 TTYDWIKLGISGSRSSIFL---AGLWWVWRHRNLMCLNNET---LSLFRLCNNIISAADA 396
+ + I I + A ++ +W RNL L ++T + + + +N IS
Sbjct: 363 NFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQRASNCISDYKQ 542
Query: 397 ANSAFNGEENPPHSDRL-------------VKWNNRNHQDFILNVDGSCIGTPPRTGYGG 443
AN+ + PP R KW N +N D + + + G G
Sbjct: 543 ANT-----QAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDAN-LQNHGKWGLGI 704
Query: 444 VLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSDS 495
++R+ GL ++ + + L AE A+ G++ D ++ D+
Sbjct: 705 IIRDEVGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDN 860
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 63.5 bits (153), Expect = 2e-10
Identities = 56/226 (24%), Positives = 103/226 (44%), Gaps = 21/226 (9%)
Frame = -3
Query: 124 LYASANNRSSPTWSSIIRAKNILKDGYTWRIGSG-SSSFWFSN--WNSLSPIGNY----- 175
L A + +S W SI A++++K G IG+G +++ W W L+ + N+
Sbjct: 762 LNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWK-LTCVTNHPNKHS 586
Query: 176 -----VPVIDVHDLQLSVRDVLSAPGNHARVLYTNLPPDVENHINNIHTQFNDNIEDALI 230
P + ++ S D +A ++ + P I +IH Q ED+
Sbjct: 585 SRAY*APTLYGYEGCRS--DDPMRRERNANLINSIFPEGTRRKILSIHPQGPIG-EDSYS 415
Query: 231 WSQNKNGTYTTKSGFNWLITSRVPAI-------EPILH-AWSWIWRLKVPEKYKFLIWLA 282
W +K+G Y+ KSG+ ++ T+ + A +P L + +W+ K + +W
Sbjct: 414 WEYSKSGHYSVKSGY-YVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRC 238
Query: 283 CQNAVPTLSLLYHRSMAPSPICTRCGEEDETFMHCVRDCCFSKDIW 328
N++PT + + R ++ C+RCG E ET H + C +++ IW
Sbjct: 237 ISNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIW 100
>AW980456
Length = 779
Score = 51.6 bits (122), Expect = 8e-07
Identities = 25/66 (37%), Positives = 35/66 (52%), Gaps = 3/66 (4%)
Frame = -2
Query: 227 DALIWSQNKNGTYTTKSGFNWLITSRVPAIEPILHA---WSWIWRLKVPEKYKFLIWLAC 283
D LIW K+G Y KS + + + + LH WS IW+LKVP K + L+W C
Sbjct: 208 DRLIWKDEKHGKYYVKSAYRFCVEELFDS--SYLHRPGNWSGIWKLKVPPKVQNLVWRMC 35
Query: 284 QNAVPT 289
+ +PT
Sbjct: 34 RGCLPT 17
>CA921218
Length = 707
Score = 50.8 bits (120), Expect = 1e-06
Identities = 35/121 (28%), Positives = 54/121 (43%)
Frame = -3
Query: 442 GGVLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSDSLLAVNL 501
GG RNS F+ F+ N + L A+L+ + +++ N + + DS L VN
Sbjct: 531 GGTFRNSNAYFLLCFAENTGNG-NALHAKLSGAMRAIEIAAARNWSHLWLELDSSLVVNA 355
Query: 502 IANDTPRYHIYAALIQNIKDLLNDRNISLHHTLREGNQCADFFAKFGANSDAALVVHQSP 561
+ + + N L++ N + H REGNQCAD A FG + D + P
Sbjct: 354 FKSKSVIHWNLRNRWNNCLFLISSMNFFVSHVFREGNQCADGLANFGLSLDHLTYWNHVP 175
Query: 562 P 562
P
Sbjct: 174 P 172
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 50.1 bits (118), Expect = 2e-06
Identities = 35/137 (25%), Positives = 56/137 (40%), Gaps = 12/137 (8%)
Frame = -2
Query: 266 IWRLKVPEKYKFLIWLACQNAVPTLSLLYHRSMAPSP--ICTRCGEEDETFMHCVRDCCF 323
IW +VP K W ++ +PT S L +R + P+ +C E+ H C +
Sbjct: 584 IWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLSCSY 405
Query: 324 SKDIWQK----IGFTSNDFVTTATTYDWIKLGISGSRSS------IFLAGLWWVWRHRNL 373
+W IGF D + + G+++S I+L W +W RN
Sbjct: 404 FASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAWVLWTERNN 225
Query: 374 MCLNNETLSLFRLCNNI 390
MC N+ L RL + +
Sbjct: 224 MCFNDSITPLPRLLDKV 174
>BG585499
Length = 792
Score = 48.9 bits (115), Expect = 5e-06
Identities = 30/125 (24%), Positives = 53/125 (42%), Gaps = 15/125 (12%)
Frame = +3
Query: 263 WSWIWRLKVPEKYKFLIWLACQNAVPTLSLLYHRSMAPSPI---CTRCGEEDETFMHCVR 319
W +W + P + + +WL + T Y RS + + C CG DET +H +
Sbjct: 225 WKMLWGWRGPHRTQTFMWLVAHGCILTN---YRRSRWGTRVLATCPCCGNADETVLHVLC 395
Query: 320 DCCFSKDIWQKIGFTSNDFVTTATTY----DWIKLGISGSRSSI--------FLAGLWWV 367
DC + +W I +D++T ++ DW+ +S + + F+ W +
Sbjct: 396 DCRPASQVW--IRLVPSDWITNFFSFDDCRDWVFKNLSKRSNGVSKFKWQPTFMTTCWHM 569
Query: 368 WRHRN 372
W RN
Sbjct: 570 WTWRN 584
>BF006335 similar to GP|14334538|gb| unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 505
Score = 43.9 bits (102), Expect(2) = 3e-05
Identities = 32/88 (36%), Positives = 48/88 (54%)
Frame = +2
Query: 439 TGYGGVLRNSAGLFISGFSGLIPNSADILLAELTAIHQGLKLVTDLNVNEIMCYSDSLLA 498
+G+GG+ RN F GF G I S +IL AE+ + GLKL + +CY DSL
Sbjct: 206 SGFGGLDRNYDKAFQFGFYGSIGWS-NILHAEIQDLLVGLKLWW--KARKFICYYDSLHV 376
Query: 499 VNLIANDTPRYHIYAALIQNIKDLLNDR 526
V L + +T +H YA +++ + L +R
Sbjct: 377 VQLGSMNTHHFHHYANMLEIYQSLHEER 460
Score = 21.9 bits (45), Expect(2) = 3e-05
Identities = 9/14 (64%), Positives = 9/14 (64%)
Frame = +3
Query: 525 DRNISLHHTLREGN 538
D N SLH T EGN
Sbjct: 459 DWNFSLHQTFPEGN 500
>TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Brassica
oleracea}, partial (3%)
Length = 1156
Score = 46.2 bits (108), Expect = 3e-05
Identities = 20/44 (45%), Positives = 29/44 (65%)
Frame = -3
Query: 527 NISLHHTLREGNQCADFFAKFGANSDAALVVHQSPPADLLPLLR 570
N++++HT RE N+C DFF KF A+S+ + H S +LL LR
Sbjct: 134 NVTIYHTFREVNRCIDFFVKFDASSNFEFLCHVSTSTNLLNFLR 3
>BG586862
Length = 804
Score = 44.7 bits (104), Expect = 1e-04
Identities = 31/120 (25%), Positives = 51/120 (41%), Gaps = 7/120 (5%)
Frame = -1
Query: 266 IWRLKVPEKYKFLIWLACQNAVPTLSLLYHRSMAPSPICTRCGEEDETFMHCVRDCCFSK 325
+W +K ++K +W NA+P L+ R + S +C RC + ET H +C ++
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 326 DIW----QKIGFTSNDFVTTATTYDWIKLGISGSRSSIFL---AGLWWVWRHRNLMCLNN 378
W I F S+ + +DWI I + + A L+ +W RN N
Sbjct: 474 KEWFGSQLGINFHSSGVL---HFHDWITNFILKNDEETIIALTALLYSIWHARNQKVFEN 304
>BG584442
Length = 775
Score = 44.3 bits (103), Expect = 1e-04
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 2/108 (1%)
Frame = +1
Query: 9 KNRLLNKTGRLTLANSVLTSIPSYFMQTNWLPQNICDSIDQTTRNFIWR--GTTNKGIHL 66
+N+ L+K + L SI SY M L + D I++ F W G KG+H
Sbjct: 397 RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGENRKGMHW 576
Query: 67 VNWKKISLPKNIGGLSIRAAREANTSLLGKLVWDMVQSSNKLWVSLLS 114
++ +K+ + KN GG+ N +LGK V + + L++ +S
Sbjct: 577 MS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLFLEKIS 720
>TC79715 similar to PIR|C86277|C86277 hypothetical protein AAF43930.1
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 42.7 bits (99), Expect = 4e-04
Identities = 18/37 (48%), Positives = 25/37 (66%)
Frame = +2
Query: 544 FAKFGANSDAALVVHQSPPADLLPLLRADALGIYFVR 580
FAK GA S+ + +PP DL+PL++ DA+G YF R
Sbjct: 1004 FAKLGATSNEEFTTYFTPPEDLIPLIKNDAMGTYFPR 1114
>TC88299 weakly similar to GP|17979137|gb|AAL49826.1 unknown protein
{Arabidopsis thaliana}, partial (36%)
Length = 1167
Score = 42.7 bits (99), Expect = 4e-04
Identities = 25/61 (40%), Positives = 35/61 (56%), Gaps = 3/61 (4%)
Frame = -3
Query: 516 IQNIKDLLNDRNISLHHTL---REGNQCADFFAKFGANSDAALVVHQSPPADLLPLLRAD 572
I+N + RN H++ REGNQC+DF K A+S+ L+ S +LL LLR+D
Sbjct: 193 IKNPSKFVMMRNKCQSHSINPFREGNQCSDFMTKLNASSNLELLRQDSSATNLLFLLRSD 14
Query: 573 A 573
A
Sbjct: 13 A 11
>TC91623 similar to PIR|D86241|D86241 protein T16B5.8 [imported] -
Arabidopsis thaliana, partial (26%)
Length = 982
Score = 42.0 bits (97), Expect = 7e-04
Identities = 25/51 (49%), Positives = 28/51 (54%)
Frame = +3
Query: 528 ISLHHTLREGNQCADFFAKFGANSDAALVVHQSPPADLLPLLRADALGIYF 578
I LH T +GNQC DF AK GA S+ LV S PLL DA+G F
Sbjct: 753 IMLHCTFGKGNQCDDFLAKMGAQSNDDLVFLDS------PLLLVDAMGTSF 887
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,737,815
Number of Sequences: 36976
Number of extensions: 416126
Number of successful extensions: 2288
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 2242
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2284
length of query: 581
length of database: 9,014,727
effective HSP length: 101
effective length of query: 480
effective length of database: 5,280,151
effective search space: 2534472480
effective search space used: 2534472480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135319.4


