

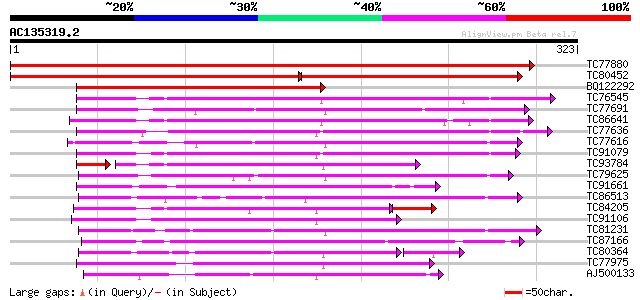
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.2 - phase: 0
(323 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77880 similar to GP|9622153|gb|AAF89645.1| seed maturation pro... 587 e-168
TC80452 similar to GP|9622153|gb|AAF89645.1| seed maturation pro... 299 8e-82
BQ122292 similar to GP|9622153|gb| seed maturation protein PM34 ... 199 1e-51
TC76545 similar to GP|6119844|gb|AAF04253.1| short-chain alcohol... 116 1e-26
TC77691 similar to SP|P28643|FABG_CUPLA 3-oxoacyl-[acyl-carrier ... 112 2e-25
TC86641 similar to GP|6119844|gb|AAF04253.1| short-chain alcohol... 106 1e-23
TC77636 similar to PIR|T11579|T11579 probable short chain alcoho... 99 2e-21
TC77616 similar to GP|11994422|dbj|BAB02424. oxidoreductase sho... 97 8e-21
TC91079 similar to PIR|T11579|T11579 probable short chain alcoho... 94 7e-20
TC93784 similar to GP|6119723|gb|AAF04193.1| short-chain alcohol... 83 8e-20
TC79625 similar to GP|6223643|gb|AAF05857.1| putative short-chai... 91 5e-19
TC91661 similar to GP|14334866|gb|AAK59611.1 unknown protein {Ar... 88 4e-18
TC86513 weakly similar to GP|15777867|gb|AAL05894.1 At1g07440/F2... 88 5e-18
TC84205 similar to PIR|T11579|T11579 probable short chain alcoho... 84 1e-17
TC91106 similar to GP|22651515|gb|AAL99237.1 short-chain dehydro... 84 6e-17
TC81231 weakly similar to GP|8978342|dbj|BAA98195.1 short chain ... 84 6e-17
TC87166 weakly similar to GP|15824408|gb|AAL09328.1 steroleosin ... 80 1e-15
TC80364 weakly similar to SP|P50165|TRNH_DATST Tropinone reducta... 71 3e-15
TC77975 weakly similar to GP|17104783|gb|AAL34280.1 putative alc... 74 6e-14
AJ500133 weakly similar to PIR|T50009|T500 hypothetical protein ... 70 1e-12
>TC77880 similar to GP|9622153|gb|AAF89645.1| seed maturation protein PM34
{Glycine max}, partial (98%)
Length = 1112
Score = 587 bits (1513), Expect = e-168
Identities = 290/299 (96%), Positives = 294/299 (97%)
Frame = +1
Query: 1 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 60
MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN
Sbjct: 79 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 258
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI 120
LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI
Sbjct: 259 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI 438
Query: 121 INAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNII 180
INAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNII
Sbjct: 439 INAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNII 618
Query: 181 NTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFN 240
NTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFN
Sbjct: 619 NTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFN 798
Query: 241 EEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINASNVIYL 299
EEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNG ++NA V+ +
Sbjct: 799 EEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGGTVVNA*MVLVI 975
>TC80452 similar to GP|9622153|gb|AAF89645.1| seed maturation protein PM34
{Glycine max}, complete
Length = 1257
Score = 299 bits (766), Expect = 8e-82
Identities = 140/167 (83%), Positives = 155/167 (91%)
Frame = +3
Query: 1 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 60
M +G QK PPQKQDTQPGKEH M+P PQFTCPDYKP+NKLQGK+AV+TGGDSGIGRAVCN
Sbjct: 72 MASGEQKFPPQKQDTQPGKEHVMDPLPQFTCPDYKPSNKLQGKVAVITGGDSGIGRAVCN 251
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI 120
LF+LEGATV FTYVKG EDKDA+DTL+ML+ AK+A+AKDPMA+ ADLGFDENCK+V+DEI
Sbjct: 252 LFSLEGATVAFTYVKGDEDKDAKDTLEMLRNAKSADAKDPMAVAADLGFDENCKKVVDEI 431
Query: 121 INAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTR 167
+NAYG IDILVNNAAEQYEC SVEEIDE RLERVFRTNIFSYFFMTR
Sbjct: 432 VNAYGHIDILVNNAAEQYECSSVEEIDESRLERVFRTNIFSYFFMTR 572
Score = 228 bits (582), Expect = 2e-60
Identities = 110/126 (87%), Positives = 118/126 (93%)
Frame = +1
Query: 167 RHALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVA 226
RHALKHMKEGS+IINTTSVNAYKGN+ L+DYTSTKGAIVAFTR LSLQLVSKGIRVNGVA
Sbjct: 712 RHALKHMKEGSSIINTTSVNAYKGNAKLLDYTSTKGAIVAFTRGLSLQLVSKGIRVNGVA 891
Query: 227 PGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPN 286
PGPIWTPLIPASF EE+TAQFG VPMKRAGQP+EVAPS+VFLASNQCSSY TGQVLHPN
Sbjct: 892 PGPIWTPLIPASFKEEETAQFGGQVPMKRAGQPIEVAPSYVFLASNQCSSYFTGQVLHPN 1071
Query: 287 GDMLIN 292
G ++N
Sbjct: 1072GGTVVN 1089
>BQ122292 similar to GP|9622153|gb| seed maturation protein PM34 {Glycine
max}, partial (48%)
Length = 437
Score = 199 bits (506), Expect = 1e-51
Identities = 98/143 (68%), Positives = 122/143 (84%), Gaps = 1/143 (0%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
KL+GK+A+VTGGDSGIGRAVC +FA EGATV FTYVKG ED+D DTL ML AKT++A+
Sbjct: 2 KLRGKVALVTGGDSGIGRAVCLIFAKEGATVAFTYVKGVEDRDKDDTLKMLLEAKTSDAQ 181
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYG-RIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
+P+AI AD+G+DENCK+V++ ++ YG ID+LVNNAAEQ+ S+EEI E +LERVFRT
Sbjct: 182 EPLAIAADIGYDENCKQVVELVVKEYGSSIDVLVNNAAEQHLRNSIEEITEQQLERVFRT 361
Query: 158 NIFSYFFMTRHALKHMKEGSNII 180
NIFS+FF+ RHALKHMKEGS+II
Sbjct: 362 NIFSHFFLVRHALKHMKEGSSII 430
>TC76545 similar to GP|6119844|gb|AAF04253.1| short-chain alcohol
dehydrogenase SAD-C {Pisum sativum}, partial (95%)
Length = 980
Score = 116 bits (290), Expect = 1e-26
Identities = 87/281 (30%), Positives = 142/281 (49%), Gaps = 8/281 (2%)
Frame = +1
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L GK+A+VTGG SGIG+ +LFA +GA ++ D +D L ++A + ++
Sbjct: 25 RLAGKVAIVTGGASGIGKETAHLFAEQGARMVVI-------ADIQDELGN-QVAASIGSR 180
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLERVFRT 157
I D+ ++ K ++ +NAYG+IDI+ +NA ++ E+D + + VF
Sbjct: 181 KCTYIHCDIANEDQVKNLVQSTVNAYGQIDIMFSNAGIASPSDQTILELDISQADHVFAV 360
Query: 158 NIFSYFFMTRHALKHMKEG---SNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
NI ++A + M EG +I+ T SV +G L DYT +K AI+ R+ S+Q
Sbjct: 361 NIRGTTLCVKYAARAMVEGRVRGSIVCTASVLGSQGVLRLTDYTISKHAIIGLMRSASVQ 540
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFN-EEKTAQFGSDVPMKRAG---QPVEVAPSFVFLA 270
L GIRVN V+P + TPL KT + + + G VA + +FL
Sbjct: 541 LAKYGIRVNCVSPNGLATPLTMKLLGASAKTVELIYEQNKRLEGVVLNTKHVADAVLFLV 720
Query: 271 SNQCSSYITGQVLHPNGDMLINASNVIYLFTHCMVFLTHSF 311
SN+ S ++TG L +G + ++ + VFL++ F
Sbjct: 721 SNE-SDFVTGLDLRVDGSYVYGKYELL*IRKKMFVFLSNLF 840
>TC77691 similar to SP|P28643|FABG_CUPLA 3-oxoacyl-[acyl-carrier protein]
reductase chloroplast precursor (EC 1.1.1.100), partial
(82%)
Length = 1481
Score = 112 bits (280), Expect = 2e-25
Identities = 77/262 (29%), Positives = 133/262 (50%), Gaps = 4/262 (1%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
K++ + +VTG GIG+A+ G V+ Y AR + + +++K A
Sbjct: 389 KVESPVVIVTGASRGIGKAIALALGKAGCKVLVNY--------ARSSKEAEEVSKEIEAL 544
Query: 99 DPMAIP--ADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
A+ D+ + + +I ++A+G ID+L+NNA + G + + + + + V
Sbjct: 545 GGQALTYGGDVSNEADVNSMIKTAVDAWGTIDVLINNAGITRD-GLLMRMKKSQWQEVID 721
Query: 157 TNIFSYFFMTRHALKHMKEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N+ F T+ A K M + IIN +SV GN+ +Y + K ++ T++++ +
Sbjct: 722 LNLTGVFLSTQAAAKIMMKNKKGRIINISSVVGLIGNAGQANYAAAKAGVIGLTKSVAKE 901
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQC 274
S+GI VN VAPG I + + A + + +P+ R GQP EVA FLA +Q
Sbjct: 902 YSSRGITVNAVAPGFIASDMT-AKLGNDLEKKILEAIPLGRYGQPEEVAGLVEFLALSQA 1078
Query: 275 SSYITGQVLHPNGDMLINASNV 296
+SYITGQV +G M++ S++
Sbjct: 1079ASYITGQVFTIDGGMVM*ISDL 1144
>TC86641 similar to GP|6119844|gb|AAF04253.1| short-chain alcohol
dehydrogenase SAD-C {Pisum sativum}, partial (93%)
Length = 1036
Score = 106 bits (265), Expect = 1e-23
Identities = 84/275 (30%), Positives = 136/275 (48%), Gaps = 11/275 (4%)
Frame = +1
Query: 35 KPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKT 94
K + +L KIA+VTGG SGIG+ ++FA +GA ++ D +D L ++A +
Sbjct: 19 KSSLRLASKIAIVTGGASGIGKETAHVFAEQGARMVVI-------ADIQDELGN-EVAAS 174
Query: 95 ANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLER 153
+ + D+ ++ K ++ +N YG++DI+ +NA +V E D + +
Sbjct: 175 IGSHRCTYVHCDVTNEDQVKNLVQSTVNTYGQVDIMFSNAGIASPSDQTVLEFDISQADH 354
Query: 154 VFRTNIFSYFFMTRHALKHMKEG---SNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRA 210
+F N+ +HA + M +G +I+ T SV G+ L DY +K AI+ R+
Sbjct: 355 LFSVNVRGMALCVKHAARAMVDGCVRGSIVCTASVAGSNGSMKLTDYVMSKHAIIGLMRS 534
Query: 211 LSLQLVSKGIRVNGVAPGPIWTPLIPASFNE-EKTAQ--FGSDVPMKRAGQPV----EVA 263
S QL GIRVN V+P + TPL + E+T FG KR V VA
Sbjct: 535 ASKQLAKHGIRVNCVSPNGLATPLTMKLLDAGEETVDLIFGE---YKRLEGVVLNTKHVA 705
Query: 264 PSFVFLASNQCSSYITGQVLHPNGDMLINASNVIY 298
+ +FL SN+ S ++TG L +G L S +++
Sbjct: 706 DAVLFLVSNE-SDFVTGLDLRVDGSYLDGKSELVF 807
>TC77636 similar to PIR|T11579|T11579 probable short chain alcohol
dehydrogenase CPRD12 drought-inducible - cowpea,
partial (84%)
Length = 1016
Score = 99.4 bits (246), Expect = 2e-21
Identities = 81/279 (29%), Positives = 132/279 (47%), Gaps = 8/279 (2%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYV---KGHEDKDARDTLDMLKMAKTA 95
+L+GK+A++TGG SGIG A LF+ GA V+ + KGH + K
Sbjct: 128 RLEGKVALITGGASGIGEATARLFSEHGAQVVIADIQDDKGHS------------ICKEL 271
Query: 96 NAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERV 154
+ D+ +E+ + ++ + YG++DI+ NNA + E E V
Sbjct: 272 QKSSSSYVRCDVTKEEDIENAVNTTVFKYGKLDIMFNNAGISGVNKTKILENKLSEFEDV 451
Query: 155 FRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRAL 211
+ N+ F T+HA + M + GS IINT SV G YTS+K A+V R
Sbjct: 452 IKVNLTGVFLGTKHAARVMIPARRGS-IINTASVGGSIGGCAPHAYTSSKHAVVGLMRNT 628
Query: 212 SLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAG-QPVEVAPSFVFLA 270
+++L GIRVN V+P + TP++ F + +K P +VA + ++L
Sbjct: 629 AIELGPFGIRVNCVSPYFLATPMVTNFFKLDDGGALDIFSNLKGTNLVPKDVAEAALYLG 808
Query: 271 SNQCSSYITGQVLHPNGDMLINASNVIYLFTHCMVFLTH 309
S++ S Y++G L +G + + A+N +F + H
Sbjct: 809 SDE-SKYVSGLNLVIDGGVSV-ANNGFCVFEQSV*IYAH 919
>TC77616 similar to GP|11994422|dbj|BAB02424. oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (98%)
Length = 1912
Score = 97.1 bits (240), Expect = 8e-21
Identities = 81/275 (29%), Positives = 138/275 (49%), Gaps = 16/275 (5%)
Frame = +3
Query: 34 YKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAK 93
+KP + L+GK+A++TGG SGIG + GA+V V G + + + +L+
Sbjct: 123 FKP-DILKGKVALITGGASGIGFEISTQLGKHGASVA---VMGRRKQVLQSAVSVLQ--- 281
Query: 94 TANAKDPMAIPA-----DLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDE 148
++IPA D+ E+ RV+D +GRIDILVN AA + + E++
Sbjct: 282 ------SLSIPAVGFEGDVRKQEDAARVVDLTFKHFGRIDILVNAAAGNF-LAAAEDLSP 440
Query: 149 PRLERVFRTNIFSYFFMTRHALKHMKEGSN---------IINTTSVNAYKGNSTLIDYTS 199
V + F M ALK++K+G+ IIN ++ Y + I ++
Sbjct: 441 NGFRTVLDIDSVGTFTMCHEALKYLKKGAPGRNSSSGGLIINISATLHYGASWYQIHVSA 620
Query: 200 TKGAIVAFTRALSLQL-VSKGIRVNGVAPGPIW-TPLIPASFNEEKTAQFGSDVPMKRAG 257
K A+ + TR L+L+ IRVNG+APGPI TP + EE ++ ++P+ + G
Sbjct: 621 AKAAVDSTTRNLALEWGTDYDIRVNGIAPGPIGETPGMSKLAPEEIGSRGRDEMPLYKLG 800
Query: 258 QPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
+ ++A + ++LAS+ YI G + +G + ++
Sbjct: 801 EKWDIAMAALYLASD-AGKYINGDTMVVDGGLWLS 902
>TC91079 similar to PIR|T11579|T11579 probable short chain alcohol
dehydrogenase CPRD12 drought-inducible - cowpea,
partial (93%)
Length = 876
Score = 94.0 bits (232), Expect = 7e-20
Identities = 73/258 (28%), Positives = 127/258 (48%), Gaps = 5/258 (1%)
Frame = +1
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A++TGG SGIG A LF+ GA V+ D +D + + + +
Sbjct: 73 RLEGKVALITGGASGIGEATARLFSNHGAQVVIA--------DIQDDIGH-SICQELHKS 225
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERVFRT 157
+ D+ +++ + ++ ++ +G++DI+ NNA ++ E + V
Sbjct: 226 SATYVHCDVTKEKDIENAVNTTVSKHGKLDIMFNNAGITGINKTNILENKLSEFQEVIDI 405
Query: 158 NIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N+ F T+HA + M + GS IINT SV G YTS+K A+V + +++
Sbjct: 406 NLTGVFLGTKHAARVMTPVRRGS-IINTASVCGCIGGVASHAYTSSKHAVVGLMKNTAIE 582
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAG-QPVEVAPSFVFLASNQ 273
L GIRVN V+P + TPL F + +K A P +VA + ++L S++
Sbjct: 583 LGPYGIRVNCVSPYVVGTPLAKNFFKLDDDGVLDVYSNLKGANLLPKDVAEAALYLGSDE 762
Query: 274 CSSYITGQVLHPNGDMLI 291
S Y++G L +G + +
Sbjct: 763 -SKYVSGHNLVVDGGLTV 813
>TC93784 similar to GP|6119723|gb|AAF04193.1| short-chain alcohol
dehydrogenase {Pisum sativum}, partial (68%)
Length = 650
Score = 82.8 bits (203), Expect(2) = 8e-20
Identities = 57/178 (32%), Positives = 94/178 (52%), Gaps = 4/178 (2%)
Frame = +2
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI 120
+FA EG V+ D +D L ++A + + I D+ ++ K ++
Sbjct: 92 VFANEGVRVVVI-------ADIQDELGN-QVAASIGIQRCTYIHCDVADEDQVKNLVRST 247
Query: 121 INAYGRIDILVNNAAEQYECG-SVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGS-- 177
++ YG++DI+ +NA +V E+D +L+R+F N+ +HA + M EGS
Sbjct: 248 VDTYGQVDIMFSNAGIVSPTDQTVMELDMSQLDRLFGVNVRGMALCVKHAARAMVEGSVR 427
Query: 178 -NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPL 234
+I+ T SV+ G+S DYT +K A++ RA S+QL + GIRVN V+P + TPL
Sbjct: 428 GSIVCTGSVSGSVGSSRSTDYTMSKHAVLGLMRAASVQLATHGIRVNCVSPNGLATPL 601
Score = 31.6 bits (70), Expect(2) = 8e-20
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = +3
Query: 39 KLQGKIAVVTGGDSGIGRA 57
+L GK+A+VTGG SGIG A
Sbjct: 24 RLSGKVAIVTGGASGIGEA 80
>TC79625 similar to GP|6223643|gb|AAF05857.1| putative short-chain type
dehydrogenase/reductase {Arabidopsis thaliana}, partial
(85%)
Length = 1039
Score = 91.3 bits (225), Expect = 5e-19
Identities = 71/258 (27%), Positives = 115/258 (44%), Gaps = 10/258 (3%)
Frame = +2
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
LQ ++A++TG GIGR + + GA ++ + + ++L +A NA
Sbjct: 56 LQNRVAIITGSSRGIGREIAIHLSSLGARIVINH-------SSSNSLSADSLAADINATS 214
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGR-----IDILVNNAA---EQYECGSVEEIDEPRL 151
P+ +G D + + + + ++ R I IL+N A + Y S+
Sbjct: 215 PLPRATVIGADISDQSQVQSLFDSAERFFNSPIHILINCAGVIDDTYP--SIANTSIESF 388
Query: 152 ERVFRTNIFSYFFMTRHALKHMKEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAFTR 209
+RVF N F R A +K G II +S YT+ K A+ T+
Sbjct: 389 DRVFGVNARGAFLCAREAANRLKRGGGGRIILFSSSQVAALRPGFAAYTAAKAAVETMTK 568
Query: 210 ALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFL 269
L+ +L GI N VAPGPI T + EE+ + + P+ R G+ +VAP FL
Sbjct: 569 ILAKELKGTGITANCVAPGPIATEMFFGGRTEEQVQKIIDESPLGRLGETKDVAPLVGFL 748
Query: 270 ASNQCSSYITGQVLHPNG 287
AS+ ++ GQ++ NG
Sbjct: 749 ASD-AGEWVNGQIIRING 799
>TC91661 similar to GP|14334866|gb|AAK59611.1 unknown protein {Arabidopsis
thaliana}, partial (98%)
Length = 1095
Score = 88.2 bits (217), Expect = 4e-18
Identities = 57/207 (27%), Positives = 107/207 (51%)
Frame = +3
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+ +GK+A+VT GIG + LEGA+V+ + K+ + L+ A
Sbjct: 96 RFKGKVAIVTASTQGIGFTIAERLGLEGASVV---ISSRRQKNVDVAAEKLR----AKGI 254
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
D A+ + K +ID+ + YG+ID++V+NAA S+ + + L++++ N
Sbjct: 255 DVFAVVCHVSNALQRKDLIDKTVQKYGKIDVVVSNAAANPSVDSILQTQDSVLDKLWEIN 434
Query: 159 IFSYFFMTRHALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSK 218
+ + + + A ++ +GS+++ +S+ Y +++ Y TK A++ T+AL+ ++ K
Sbjct: 435 VKATILLLKDAAPYLPKGSSVVIISSIAGYHPPASMAMYGVTKTALLGLTKALAGEMAPK 614
Query: 219 GIRVNGVAPGPIWTPLIPASFNEEKTA 245
RVN VAPG + P ASF +A
Sbjct: 615 -TRVNCVAPG--FVPTNFASFITSNSA 686
>TC86513 weakly similar to GP|15777867|gb|AAL05894.1 At1g07440/F22G5_16
{Arabidopsis thaliana}, partial (89%)
Length = 1484
Score = 87.8 bits (216), Expect = 5e-18
Identities = 75/258 (29%), Positives = 122/258 (47%), Gaps = 5/258 (1%)
Frame = +1
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLD--MLKMAKTANA 97
L+G A+VTGG GIG + A GATV + + + + L+ + K K +
Sbjct: 55 LKGTTALVTGGSKGIGYDIVEQLAELGATV---HTCARNEAELNECLNQWVTKGYKITGS 225
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
+A A ++ RV E G+++ILVNN + +++ E + T
Sbjct: 226 VCDVASRAQR--EDLIARVSSEF---NGKLNILVNNVGTNMQKQTLD-FTEQDFSFLVNT 387
Query: 158 NIFSYFFMTR--HALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
N+ S F +++ H L ++I+ +S+ + Y++ KGAI+ T+ L+ +
Sbjct: 388 NLESAFHISQLAHPLLKASNNASIVFMSSIGGVASLNIGTIYSAAKGAIIQLTKNLACEW 567
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFNEEKTAQ-FGSDVPMKRAGQPVEVAPSFVFLASNQC 274
IR N VAPGPI TPL +EK F P+ R G+P EV+ FL
Sbjct: 568 AKDNIRTNCVAPGPIRTPLAAEHLKDEKLLDAFIERTPLGRIGEPEEVSSLVAFLCL-PA 744
Query: 275 SSYITGQVLHPNGDMLIN 292
+S+ITGQ + +G + +N
Sbjct: 745 ASFITGQTICIDGGLTVN 798
>TC84205 similar to PIR|T11579|T11579 probable short chain alcohol
dehydrogenase CPRD12 drought-inducible - cowpea,
partial (61%)
Length = 645
Score = 84.3 bits (207), Expect(2) = 1e-17
Identities = 56/187 (29%), Positives = 90/187 (47%), Gaps = 4/187 (2%)
Frame = +1
Query: 37 ANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTAN 96
A + +GK+A++TGG SGIG A F GA VI D +D L + K
Sbjct: 7 AKRFEGKVAIITGGASGIGAATAKPFVQYGAKVIIA--------DIQDELGQ-SLCKNLG 159
Query: 97 AKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA--EQYECGSVEEIDEPRLERV 154
K+ + + D+ + + K V+D ++ YG++DI+ NNA + + D +RV
Sbjct: 160 TKNILCVHCDVTIESDIKTVVDIAVSNYGKLDIMFNNAGI*SDDKNREILNYDSEAFKRV 339
Query: 155 FRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALS 212
F N++ F +HA + M ++ I+ T SV T YT +K A+V T+ L
Sbjct: 340 FDVNVYGAFLGAKHAARVMIPQKKGVILFTASVATKIAGETTHAYTCSKHALVGLTKNLC 519
Query: 213 LQLVSKG 219
++L G
Sbjct: 520 VELGKYG 540
Score = 22.3 bits (46), Expect(2) = 1e-17
Identities = 10/25 (40%), Positives = 15/25 (60%)
Frame = +2
Query: 219 GIRVNGVAPGPIWTPLIPASFNEEK 243
GIRVN ++P + TP++ S K
Sbjct: 539 GIRVNCISPPALPTPILMNSLKLNK 613
>TC91106 similar to GP|22651515|gb|AAL99237.1 short-chain
dehydrogenase/reductase {Arabidopsis thaliana}, partial
(32%)
Length = 627
Score = 84.3 bits (207), Expect = 6e-17
Identities = 54/191 (28%), Positives = 92/191 (47%), Gaps = 3/191 (1%)
Frame = +3
Query: 36 PANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTA 95
PA +L GK+AVVTGG SGIG ++ LF GA V D +D L +
Sbjct: 78 PAQRLLGKVAVVTGGASGIGESIVRLFHTHGAKVCIA--------DIQDDLGQKLFDSLS 233
Query: 96 NAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERV 154
+ ++ + D+ + + + + +G +DI+VNNA C + +D ++V
Sbjct: 234 DLENVFFVHCDVAVEADVSTAVSIAVAKFGTLDIMVNNAGISGAPCPDIRNVDMAEFDKV 413
Query: 155 FRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALS 212
F N+ F +HA +++ K+ +II+ +SV + YT +K A+ T ++
Sbjct: 414 FNINVKGVFHGMKHAAQYLIPKKSGSIISISSVASSLXGLGPHGYTGSKHAVWGLTXNVA 593
Query: 213 LQLVSKGIRVN 223
+L + IRVN
Sbjct: 594 XELGNHXIRVN 626
>TC81231 weakly similar to GP|8978342|dbj|BAA98195.1 short chain alcohol
dehydrogenase-like {Arabidopsis thaliana}, partial (95%)
Length = 1040
Score = 84.3 bits (207), Expect = 6e-17
Identities = 79/268 (29%), Positives = 123/268 (45%), Gaps = 4/268 (1%)
Frame = +2
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+G A+VTGG GIG A+ A GATV +T + E+ + R L K
Sbjct: 80 LKGFTALVTGGTRGIGHAIVEELAEFGATV-YTCSRNQEELNKR-----LNEWKEKGFSV 241
Query: 100 PMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
++ D + +I + +++ G+++I VNNA ++E E +V TN
Sbjct: 242 YGSV-CDASSSSQREELIQNVASSFNGKLNIFVNNAGTNVRKPTIEYTAED-YSKVMTTN 415
Query: 159 IFSYFFMTRHALKHMKEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
+ S + + + +KE N I+ +SV + T Y ++K AI T++L+ +
Sbjct: 416 LDSAYHLCQLTYPLLKESGNGSIVFISSVGSLTSVGTGSIYAASKAAINQLTKSLACEWA 595
Query: 217 SKGIRVNGVAPGPIWTPLIPASF-NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
IR N VAP TPL+ NEE Q S P+KR + EV+ FL +
Sbjct: 596 KDNIRSNCVAPWYTKTPLVEHLIANEEFVNQVLSRTPIKRIAETHEVSSLVTFLCL-PAA 772
Query: 276 SYITGQVLHPNGDMLINASNVIYLFTHC 303
SYITGQ++ +G +N T C
Sbjct: 773 SYITGQIVSGDGRFTVNGFQPSMRIT*C 856
>TC87166 weakly similar to GP|15824408|gb|AAL09328.1 steroleosin {Sesamum
indicum}, partial (83%)
Length = 1166
Score = 79.7 bits (195), Expect = 1e-15
Identities = 67/253 (26%), Positives = 113/253 (44%), Gaps = 1/253 (0%)
Frame = +1
Query: 42 GKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPM 101
GK+ +TG SGIG + +A GA + + D R+ D A+ + D +
Sbjct: 145 GKVVHITGASSGIGEHLAYEYAKRGARLALS---ARRDTALREVADR---ARDCGSPDVI 306
Query: 102 AIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFS 161
+ AD+ ++C+R++DE +N +GR+D LVNNAA D + TN +
Sbjct: 307 IMRADVSKVDDCRRLVDETVNHFGRLDHLVNNAAISAAMMFEGVTDITNWRPLMDTNFWG 486
Query: 162 YFFMTRHALKHMKEG-SNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGI 220
+ TR AL H++ I+ +S++++ Y ++K A+V+ L ++ V +
Sbjct: 487 SVYTTRFALTHLRNSRGKIVVLSSIDSWMPAPRRSIYNASKAALVSLYETLRVE-VGADV 663
Query: 221 RVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITG 280
V V PG I + L + G D M+ VEV+ + V S S + G
Sbjct: 664 GVTIVTPGYIESELTKGKVLLPPEGKMGVDQDMR----DVEVSATPVGSVSECAKSIVNG 831
Query: 281 QVLHPNGDMLINA 293
+ GD + A
Sbjct: 832 TL---RGDRYLTA 861
>TC80364 weakly similar to SP|P50165|TRNH_DATST Tropinone reductase homolog
(EC 1.1.1.-) (P29X). [Jimsonweed Common thornapple]
{Datura stramonium}, partial (66%)
Length = 811
Score = 71.2 bits (173), Expect(2) = 3e-15
Identities = 59/187 (31%), Positives = 93/187 (49%), Gaps = 3/187 (1%)
Frame = +2
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L G A+VTGG GIG A+ A GA V +V ++D L+ K K N
Sbjct: 95 LHGMTALVTGGTRGIGYAIVEELAEFGAAV---HVCARNEEDINKCLEEWKN-KGFNVTG 262
Query: 100 PMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+ D+ F E KR+++ + + + G+++ILVNNAA+ ++ DE + TN
Sbjct: 263 SVC---DILFHEQRKRLMETVSSIFHGKLNILVNNAAKPTSKKIIDNTDED-INTTLGTN 430
Query: 159 IFSYFFMTRHALKHMKEGS--NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
S + + + A +K+ +I+ T+SV K L Y +TKGA+ FT+ L+L+
Sbjct: 431 FVSGYHLCQLAHPLLKQSGYGSIVFTSSVAGLKAIPVLSVYAATKGAVNQFTKNLALEWA 610
Query: 217 SKGIRVN 223
IR N
Sbjct: 611 KDNIRAN 631
Score = 27.7 bits (60), Expect(2) = 3e-15
Identities = 15/39 (38%), Positives = 20/39 (50%), Gaps = 4/39 (10%)
Frame = +3
Query: 225 VAPGPIWTPLIPASFN----EEKTAQFGSDVPMKRAGQP 259
VAPGP+ T L+ + N +E S PM R G+P
Sbjct: 636 VAPGPVKTSLLQSITNDNEGDEAVXGVXSQTPMGRMGEP 752
>TC77975 weakly similar to GP|17104783|gb|AAL34280.1 putative alcohol
dehydrogenase {Arabidopsis thaliana}, partial (82%)
Length = 721
Score = 74.3 bits (181), Expect = 6e-14
Identities = 55/208 (26%), Positives = 93/208 (44%), Gaps = 4/208 (1%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A+VTGG SGIG F GA V+ + ++ + K++
Sbjct: 74 RLEGKVAIVTGGASGIGAETAKTFVENGAFVVIADINDELGHQVATSIGLDKVSYHH--- 244
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERVFRT 157
D+ ++ + + + YG +DI+ +NA E S+ E D +
Sbjct: 245 ------CDVRDEKQVEETVAFALEKYGTLDIMFSNAGIEGGMSSSILEFDLNEFDNTMAI 406
Query: 158 NIFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N+ +HA + M E +II T SV A + DY ++K ++ R+ +
Sbjct: 407 NVRGSLAAIKHAARFMVERKIRGSIICTASVAASVAGNRGHDYVTSKHGLLGLVRSTCGE 586
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEE 242
L + GIRVN ++P + TPL + N E
Sbjct: 587 LGAYGIRVNSISPYGVATPLACRALNME 670
>AJ500133 weakly similar to PIR|T50009|T500 hypothetical protein T31P16.40 -
Arabidopsis thaliana, partial (32%)
Length = 623
Score = 70.1 bits (170), Expect = 1e-12
Identities = 62/212 (29%), Positives = 95/212 (44%), Gaps = 7/212 (3%)
Frame = +3
Query: 43 KIAVVTG-GDSGIGRAVCNLFALEGATVIFT----YVKGHEDKDARDTLDMLKMAKTANA 97
+I +VTG GIG C FA + VI + +K D ++ + ++ L++
Sbjct: 36 EIVLVTGCAKGGIGYEYCKAFAEKNCRVIASDISSRIKDMSDFESDNNIETLEL------ 197
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
D+ D++ +D II+ YGRIDILVNNA G + E+ + +
Sbjct: 198 --------DVSSDQSATSAVDTIISKYGRIDILVNNAGIG-STGPLAELPLDTIRKTLEI 350
Query: 158 NIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
N M + + HM K+ I+N SV Y ++K AI A + +L L+L
Sbjct: 351 NTLGQLRMVQQVVPHMALKKSGTIVNVGSVVGNISTPWAGSYCASKSAIHAMSNSLRLEL 530
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFNEEKTAQF 247
GI V V PG I + L A N EK + +
Sbjct: 531 KPFGINVVLVMPGSIRSNLGKA--NLEKLSDY 620
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,337,831
Number of Sequences: 36976
Number of extensions: 125180
Number of successful extensions: 835
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 783
length of query: 323
length of database: 9,014,727
effective HSP length: 96
effective length of query: 227
effective length of database: 5,465,031
effective search space: 1240562037
effective search space used: 1240562037
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135319.2


