

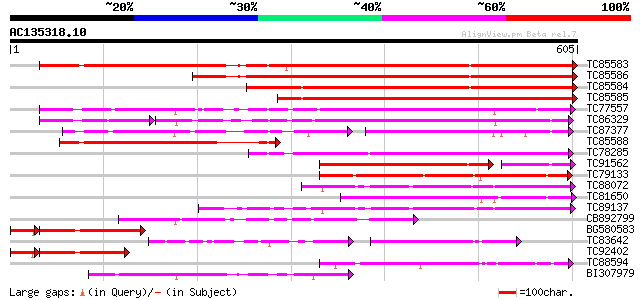
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135318.10 + phase: 0 /pseudo/partial
(605 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase ... 760 0.0
TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin... 582 e-166
TC85584 weakly similar to PIR|A84473|A84473 probable serine prot... 555 e-158
TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin... 484 e-137
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 404 e-113
TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 270 6e-94
TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like prot... 177 1e-89
TC85588 similar to GP|20198169|gb|AAM15440.1 subtilisin-like ser... 243 1e-64
TC78285 similar to PIR|A84473|A84473 probable serine proteinase ... 240 1e-63
TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase ... 186 2e-54
TC79133 similar to GP|14150446|gb|AAK53065.1 subtilisin-type pro... 204 8e-53
TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase ... 192 4e-49
TC81650 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 191 5e-49
TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycop... 191 5e-49
CB892799 weakly similar to PIR|T05768|T05 subtilisin-like protei... 183 2e-46
BG580583 weakly similar to GP|9757901|dbj serine protease-like p... 162 2e-46
TC83642 similar to PIR|S52769|S52769 subtilisin-like proteinase ... 129 4e-43
TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like ser... 148 3e-41
TC88594 similar to PIR|T08978|T08978 serine proteinase homolog F... 165 4e-41
BI307979 weakly similar to GP|20521377|db putative subtilisin-li... 164 7e-41
>TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase AIR3
auxin-induced [imported] - Arabidopsis thaliana
(fragment), partial (40%)
Length = 2691
Score = 760 bits (1962), Expect = 0.0
Identities = 401/591 (67%), Positives = 457/591 (76%), Gaps = 17/591 (2%)
Frame = +2
Query: 32 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 91
TAGGNFVP AS+F IGNGT+KGGSP++RV TYKVCWS + C+GADVL+AIDQA
Sbjct: 722 TAGGNFVPDASVFAIGNGTVKGGSPRARVATYKVCWSLLDLED----CFGADVLAAIDQA 889
Query: 92 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 151
ISDGVDIIS+S+ G S E+IFTDE+SIGAF A ++NILLVASAGN GPT GSV NVA
Sbjct: 890 ISDGVDIISLSLAGHSLVYPEDIFTDEVSIGAFHALSRNILLVASAGNEGPTGGSVVNVA 1069
Query: 152 PWVFTVAASTIDRDFSSTITIGNKT--GASLFVNLPPNQSFTLVDSIDAKFANVTNQDAR 209
PWVFT+AAST+DRDFSSTITIGN+T GASLFVNLPPNQ+F L+ S D K AN TN DA+
Sbjct: 1070 PWVFTIAASTLDRDFSSTITIGNQTIRGASLFVNLPPNQAFPLIVSTDGKLANATNHDAQ 1249
Query: 210 FCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMI 269
FCKPGTLDPSKV GKIVEC+ E G + SV++G+EALSAGAKGM+
Sbjct: 1250 FCKPGTLDPSKVKGKIVECIRE------------GNI-----KSVAEGQEALSAGAKGML 1378
Query: 270 LRNQPKFNGKTLLAESNVLSTINY--------------YDKHQLTRGHSIGISTTDT-IK 314
L NQPK GKT LAE + LS + + + + I++ D+ +K
Sbjct: 1379 LSNQPK-QGKTTLAEPHTLSCVEVPHHAPKPPKPKKSAEQERAGSHAPAFDITSMDSKLK 1555
Query: 315 SVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVS 374
+ I+ S KT Y RKPAPVMASFSSRGPN++QP ILKPDVTAPGVNILAAYSL+AS S
Sbjct: 1556 AGTTIKFSGAKTLYGRKPAPVMASFSSRGPNKIQPSILKPDVTAPGVNILAAYSLYASAS 1735
Query: 375 NLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNK 434
NL TDNR FPFN+ QGTSMSCPHVAG AGLIKTLHPNWSPAAIKSAIMTTAT DNTN+
Sbjct: 1736 NLKTDNRNNFPFNVLQGTSMSCPHVAGIAGLIKTLHPNWSPAAIKSAIMTTATTLDNTNR 1915
Query: 435 LIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNP 494
I+DA + LA PF YGSGH+QP+ A+DPGLVYDL + DYLNFLCA GY+Q+LIS LN
Sbjct: 1916 PIQDAFENKLAIPFDYGSGHVQPDLAIDPGLVYDLGIKDYLNFLCAYGYNQQLISA-LNF 2092
Query: 495 NMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLPGYNIVVVP 554
N TF CSG HSI D NYPSITLPNL LNAVNVTR VTNVGPP TY AK QL GY IVV+P
Sbjct: 2093 NGTFICSGSHSITDFNYPSITLPNLKLNAVNVTRTVTNVGPPGTYSAKAQLLGYKIVVLP 2272
Query: 555 DSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 605
+SLTFKK GEKK FQVIVQA +VTPRG+YQFG LQWT+GKHIVRSP+TV+R
Sbjct: 2273 NSLTFKKTGEKKTFQVIVQATNVTPRGKYQFGNLQWTDGKHIVRSPITVRR 2425
>TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin-like
protease {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 1460
Score = 582 bits (1500), Expect = e-166
Identities = 301/411 (73%), Positives = 334/411 (81%), Gaps = 1/411 (0%)
Frame = +1
Query: 196 IDAKFANVTNQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVS 255
IDAKF+N T +DARFC+P TLDPSKV GKIV C E G++ SV+
Sbjct: 1 IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACARE------------GKI-----KSVA 129
Query: 256 QGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKS 315
+G+EALSAGAKGM L NQPK +G TLL+E +VLST+ + +T +G++ TDTI+S
Sbjct: 130 EGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIES 309
Query: 316 VIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSN 375
KIR SQ T RKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFAS SN
Sbjct: 310 GTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASN 489
Query: 376 LVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKL 435
L+TDNRRGFPFN+ QGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTAT RDNTNK
Sbjct: 490 LLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNTNKP 669
Query: 436 IRDAIDKTLANPFAYGSG-HIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNP 494
I DA DKTLA+PFAYGSG + P +A+DPGLVYDL + DYLNFLCA+GY+++LIS LN
Sbjct: 670 ISDAFDKTLADPFAYGSGTYSHPTSAIDPGLVYDLGIKDYLNFLCASGYNKQLISA-LNF 846
Query: 495 NMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLPGYNIVVVP 554
NMTFTCSG HSI+DLNYPSITLPNLGLNA+ VTR VTNVGPPSTYFAKVQLPGY I VVP
Sbjct: 847 NMTFTCSGTHSIDDLNYPSITLPNLGLNAITVTRTVTNVGPPSTYFAKVQLPGYKIAVVP 1026
Query: 555 DSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 605
SL FKK GEKK FQVIVQA S PR +YQFGEL+WTNGKHIVRSPVTVQR
Sbjct: 1027SSLNFKKIGEKKTFQVIVQATSEIPRRKYQFGELRWTNGKHIVRSPVTVQR 1179
>TC85584 weakly similar to PIR|A84473|A84473 probable serine proteinase
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1241
Score = 555 bits (1430), Expect = e-158
Identities = 282/353 (79%), Positives = 308/353 (86%)
Frame = +1
Query: 253 SVSQGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDT 312
SV++G+EALSAGAKG+ILRNQP+ NGKTLL+E +VLSTI+Y H T G S+ I +D
Sbjct: 10 SVAEGQEALSAGAKGVILRNQPEINGKTLLSEPHVLSTISYPGNHSRTTGRSLDIIPSD- 186
Query: 313 IKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFAS 372
IKS K+RMS KT RRKPAPVMAS+SSRGPN+VQP ILKPDVTAPGVNILAAYSLFAS
Sbjct: 187 IKSGTKLRMSPAKTLNRRKPAPVMASYSSRGPNKVQPSILKPDVTAPGVNILAAYSLFAS 366
Query: 373 VSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNT 432
SNL+TD RRGFPFN+ QGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTAT RDNT
Sbjct: 367 ASNLITDTRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNT 546
Query: 433 NKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLL 492
NK I DA DKTLANPFAYGSGHI+PN+AMDPGLVYDL + DYLNFLCA+GY+Q+LIS L
Sbjct: 547 NKPISDAFDKTLANPFAYGSGHIRPNSAMDPGLVYDLGIKDYLNFLCASGYNQQLISA-L 723
Query: 493 NPNMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLPGYNIVV 552
N NMTFTCSG SI+DLNYPSITLPNLGLN+V VTR VTNVGPPSTYFAKVQL GY I V
Sbjct: 724 NFNMTFTCSGTSSIDDLNYPSITLPNLGLNSVTVTRTVTNVGPPSTYFAKVQLAGYKIAV 903
Query: 553 VPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 605
VP SL FKK GEKK FQVIVQA SVTPR +YQFGEL+WTNGKHIVRSPVTV+R
Sbjct: 904 VPSSLNFKKIGEKKTFQVIVQATSVTPRRKYQFGELRWTNGKHIVRSPVTVRR 1062
>TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin-like
serine protease {Oryza sativa (japonica
cultivar-group)}, partial (21%)
Length = 1353
Score = 484 bits (1247), Expect = e-137
Identities = 246/320 (76%), Positives = 270/320 (83%)
Frame = +2
Query: 286 NVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPN 345
+V+STINY+D +T I+ D IK+ IRMS RKPAPVMASFSSRGPN
Sbjct: 2 SVVSTINYHDPRSITTPKGSEITPED-IKTNATIRMSPANALNGRKPAPVMASFSSRGPN 178
Query: 346 QVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGL 405
+VQPYILKPDVTAPGVNILAAYSL ASVSNLVTDNRRGFPFNIQQGTSMSCPHV GTAGL
Sbjct: 179 KVQPYILKPDVTAPGVNILAAYSLLASVSNLVTDNRRGFPFNIQQGTSMSCPHVVGTAGL 358
Query: 406 IKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGL 465
IKTLHPNWSPAAIKSAIMTTAT RDNTN+ I DA + T AN FAYGSGHIQPN+A+DPGL
Sbjct: 359 IKTLHPNWSPAAIKSAIMTTATTRDNTNEPIEDAFENTTANAFAYGSGHIQPNSAIDPGL 538
Query: 466 VYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLNYPSITLPNLGLNAVN 525
VYDL + DYLNFLCAAGY+Q+LIS+L+ NMTFTC G SINDLNYPSITLPNLGLNAV
Sbjct: 539 VYDLGIKDYLNFLCAAGYNQKLISSLIF-NMTFTCYGTQSINDLNYPSITLPNLGLNAVT 715
Query: 526 VTRIVTNVGPPSTYFAKVQLPGYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQF 585
VTR VTNVGP STY AK QLPGY IVVVP SL FKK GEKK F+V VQA SVTP+G+Y+F
Sbjct: 716 VTRTVTNVGPRSTYTAKAQLPGYKIVVVPSSLKFKKIGEKKTFKVTVQATSVTPQGKYEF 895
Query: 586 GELQWTNGKHIVRSPVTVQR 605
GELQW+NGKHIVRSP+T++R
Sbjct: 896 GELQWSNGKHIVRSPITLRR 955
Score = 37.7 bits (86), Expect = 0.013
Identities = 14/20 (70%), Positives = 18/20 (90%)
Frame = +1
Query: 580 RGRYQFGELQWTNGKHIVRS 599
+G+Y+FGEL WTNGKH V+S
Sbjct: 952 KGKYEFGEL*WTNGKHCVKS 1011
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 404 bits (1039), Expect = e-113
Identities = 243/581 (41%), Positives = 336/581 (57%), Gaps = 9/581 (1%)
Frame = +3
Query: 32 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 91
TA G+ VP AS+FG +GT +G + ++RV YKVCW C+ +D+L+AID+A
Sbjct: 888 TAAGSVVPDASLFGYASGTARGMATRARVAVYKVCWK--------GGCFSSDILAAIDKA 1043
Query: 92 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 151
ISD V+++S+S+GG S + F D ++IGAF A K IL+ SAGN GP+ S++NVA
Sbjct: 1044 ISDNVNVLSLSLGGGMS----DYFRDSVAIGAFSAMEKGILVSCSAGNAGPSAYSLSNVA 1211
Query: 152 PWVFTVAASTIDRDFSSTITIGNK---TGASLFV-NLPPNQSFTLVDSIDAKFANVTNQD 207
PW+ TV A T+DRDF +++++GN +G SL+ N P L+ + +A N TN
Sbjct: 1212 PWITTVGAGTLDRDFPASVSLGNGLNYSGVSLYRGNALPESPLPLIYAGNA--TNATN-- 1379
Query: 208 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 267
C GTL P V+GKIV C ++ R V +G +AG G
Sbjct: 1380 GNLCMTGTLSPELVAGKIVLC----------DRGMNAR--------VQKGAVVKAAGGLG 1505
Query: 268 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTS 327
M+L N NG+ L+A++++L ++ G++I K +KI K
Sbjct: 1506 MVLSNTAA-NGEELVADTHLLPATAVGERE----GNAIKKYLFSEAKPTVKIVFQGTKVG 1670
Query: 328 YRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFN 387
+P+PV+A+FSSRGPN + P ILKPD+ APGVNILA +S + L D RR FN
Sbjct: 1671 V--EPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLAVDERR-VDFN 1841
Query: 388 IQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANP 447
I GTSMSCPHV+G A LIK+ HP+WSPAA++SA+MTTA I ++D+ + P
Sbjct: 1842 IISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYIAYKNGNKLQDSATGKSSTP 2021
Query: 448 FAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSIN 507
F +GSGH+ P A++PGLVYDL+ DYL FLCA Y+ I++L +S++
Sbjct: 2022 FDHGSGHVDPVAALNPGLVYDLTADDYLGFLCALNYTATQITSLARRKFQCDAGKKYSVS 2201
Query: 508 DLNYPSITL---PNLGLNAVNVTRIVTNVGPPSTYFAKVQLPGYN--IVVVPDSLTFKKN 562
DLNYPS + G N V TRI TNVGP TY A V N I V P+ L+FK N
Sbjct: 2202 DLNYPSFAVVFDTMGGANVVKHTRIFTNVGPAGTYKASVTSDSKNVKITVEPEELSFKAN 2381
Query: 563 GEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 603
EKK F V + TP+ FG L+WTNGK++V SP+++
Sbjct: 2382 -EKKSFTVTFTSSGSTPQKLNGFGRLEWTNGKNVVGSPISI 2501
>TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC 3.4.21.-)
1 - tomato, partial (91%)
Length = 2671
Score = 270 bits (690), Expect(2) = 6e-94
Identities = 187/462 (40%), Positives = 242/462 (51%), Gaps = 16/462 (3%)
Frame = +3
Query: 156 TVAASTIDRDFSSTITIGNKT---GASLFVN-LPPNQSFTLVDSIDAKFANVTNQDA-RF 210
TV A TIDRDF + IT+GN G SL+ LPPN LV + ANV+ +
Sbjct: 1086 TVGARTIDRDFPAYITLGNGNRYNGVSLYNGKLPPNSPLPLVYA-----ANVSQDSSDNL 1250
Query: 211 CKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMIL 270
C +L PSKVSGKIV C P + + L AG GMIL
Sbjct: 1251 CSTDSLIPSKVSGKIVIC-------DRGGNPRAEKSL-----------VVKRAGGIGMIL 1376
Query: 271 RNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRR 330
N + G+ L+A+S +L +K + I + KI T +
Sbjct: 1377 ANNQDY-GEELVADSFLLPAAALGEK----ASNEIKKYASSAPNPTAKIAFGG--TRFGV 1535
Query: 331 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQ 390
+P+PV+A+FSSRGPN + P ILKPD+ APGVNILA +S + L D R FNI
Sbjct: 1536 QPSPVVAAFSSRGPNILTPKILKPDLIAPGVNILAGWSGKVGPTGLSVDTRH-VSFNIIS 1712
Query: 391 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAY 450
GTSMSCPHV+G A L+K HP WSPAAI+SA+MTT+ + I+D A P Y
Sbjct: 1713 GTSMSCPHVSGLAALLKGAHPEWSPAAIRSALMTTSYGTYKNGQTIKDVATGIPATPLDY 1892
Query: 451 GSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLN 510
GSGH+ P A+DPGLVYD + DYLNFLCA Y+ I + T + + DLN
Sbjct: 1893 GSGHVDPVAALDPGLVYDATTDDYLNFLCALNYNSFQIKLVARREFTCDKRIKYRVEDLN 2072
Query: 511 YPSITLP---------NLGLNAVNVTRIVTNVGPPSTYFAKV--QLPGYNIVVVPDSLTF 559
YPS ++P + + V RI+TNVG PSTY V Q P IVV P +L+F
Sbjct: 2073 YPSFSVPFDTASGRGSSHNPSIVQYKRILTNVGAPSTYKVSVSSQSPLDKIVVEPQTLSF 2252
Query: 560 KKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPV 601
K+ EKK + V + S+ P G F L+W++GKH V SP+
Sbjct: 2253 KELNEKKSYTVTFTSHSM-PSGTTSFAHLEWSDGKHKVTSPI 2375
Score = 93.2 bits (230), Expect(2) = 6e-94
Identities = 48/123 (39%), Positives = 70/123 (56%)
Frame = +2
Query: 32 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 91
TA G+ V GAS+FG +GT KG + ++RV YKVCW C+ +D+ +AID+A
Sbjct: 740 TAAGSAVAGASLFGFASGTAKGMATQARVAAYKVCWL--------GGCFTSDIAAAIDKA 895
Query: 92 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 151
I + V R + + D +++G F A IL+ +SAGNGGP+ S+ NVA
Sbjct: 896 IDTQARTAAYKVS-RLGGGLTDYYKDTVAMGTFAAIEHGILVSSSAGNGGPSKASLANVA 1072
Query: 152 PWV 154
PW+
Sbjct: 1073PWI 1081
>TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (54%)
Length = 2071
Score = 177 bits (449), Expect(2) = 1e-89
Identities = 119/319 (37%), Positives = 173/319 (53%), Gaps = 10/319 (3%)
Frame = +2
Query: 57 KSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFT 116
++RV YKVCW C+ +D+ + +D+AI DGV+I+S+S+GG + + +
Sbjct: 8 EARVAAYKVCWLSG--------CFTSDIAAGMDKAIEDGVNILSMSIGG----SIMDYYR 151
Query: 117 DEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITIGN-- 174
D I+IGAF A + IL+ +SAGNGGP+ S++NVAPW+ TV A TIDRDF S IT+GN
Sbjct: 152 DIIAIGAFTAMSHGILVSSSAGNGGPSAESLSNVAPWITTVGAGTIDRDFPSYITLGNGK 331
Query: 175 -KTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARF-CKPGTLDPSKVSGKIVECVGEK 232
TGASL+ P + S V NV+ + C P +L SKV GKIV C
Sbjct: 332 TYTGASLYNGKPSSDSLLPV----VYAGNVSESSVGYLCIPDSLTSSKVLGKIVICERG- 496
Query: 233 ITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTIN 292
+ V +G +AG GMIL N + G+ L+A+S++L
Sbjct: 497 -----------------GNSRVEKGLVVKNAGGVGMILVNNEAY-GEELIADSHLLPAA- 619
Query: 293 YYDKHQLTRGHSIGISTTDTIKSVI------KIRMSQPKTSYRRKPAPVMASFSSRGPNQ 346
++G ++ +K + + ++ T + +P+PV+A+FSSRGPN
Sbjct: 620 -----------ALGQKSSTVLKDYVFTTKNPRAKLVFGGTHLQVQPSPVVAAFSSRGPNS 766
Query: 347 VQPYILKPDVTAPGVNILA 365
+ P ILKPD+ APGVNILA
Sbjct: 767 LTPKILKPDLIAPGVNILA 823
Score = 171 bits (433), Expect(2) = 1e-89
Identities = 97/237 (40%), Positives = 132/237 (54%), Gaps = 15/237 (6%)
Frame = +1
Query: 380 NRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDA 439
++R FNI GTSMSCPH +G A ++K +P WSPAAI+SA+MTTA + I D
Sbjct: 865 DKRHVNFNIISGTSMSCPHASGLAAIVKGAYPEWSPAAIRSALMTTAYTSYKNGQTIVDV 1044
Query: 440 IDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFT 499
A PF +GSGH+ P +A+DPGLVYD++V DYL F CA Y+ I T
Sbjct: 1045 ATGKPATPFDFGSGHVDPVSALDPGLVYDINVDDYLGFFCALNYTSYQIKLAARREFTCD 1224
Query: 500 CSGIHSINDLNYPSI-----TLPNLGLNA-----VNVTRIVTNVGPPSTYFAKVQLPGYN 549
+ + D NYPS T +G + V R++TNVG P TY A V L +
Sbjct: 1225 ARKKYRVEDFNYPSFAVALETASGIGGGSNKPIIVEYNRVLTNVGAPGTYNATVVLSSVD 1404
Query: 550 -----IVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPV 601
+VV P++++FK+ EKK ++V S+ P G FG L+W +GKH V SP+
Sbjct: 1405 SSSVKVVVEPETISFKEVYEKKGYKVRFICGSM-PSGTKSFGYLEWNDGKHKVGSPI 1572
>TC85588 similar to GP|20198169|gb|AAM15440.1 subtilisin-like serine
protease AIR3 {Arabidopsis thaliana}, partial (7%)
Length = 572
Score = 243 bits (621), Expect = 1e-64
Identities = 138/238 (57%), Positives = 157/238 (64%), Gaps = 2/238 (0%)
Frame = +1
Query: 54 GSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEE 113
GSP++RV TYKVCWS + C+GADVL+AIDQAISDGVDIIS+S+ G S E+
Sbjct: 1 GSPRARVATYKVCWSLLDLED----CFGADVLAAIDQAISDGVDIISLSLAGHSLVYPED 168
Query: 114 IFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITIG 173
IFTDE+SIGAF A ++NILLVASAGN GPT GSV NV PWVFT+AAST+DRDFSSTITIG
Sbjct: 169 IFTDEVSIGAFHALSRNILLVASAGNEGPTGGSVVNVTPWVFTIAASTLDRDFSSTITIG 348
Query: 174 NKT--GASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSKVSGKIVECVGE 231
N+T GASLFVNLPPNQ+F L+ S D K AN TN DA+FCKPGTLDPSKV
Sbjct: 349 NQTIRGASLFVNLPPNQAFPLIVSTDGKLANATNHDAQFCKPGTLDPSKV---------- 498
Query: 232 KITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAESNVLS 289
KGM+L NQPK GKT LAE + LS
Sbjct: 499 ----------------------------------KGMLLSNQPK-QGKTTLAEPHTLS 567
>TC78285 similar to PIR|A84473|A84473 probable serine proteinase [imported]
- Arabidopsis thaliana, partial (41%)
Length = 1353
Score = 240 bits (612), Expect = 1e-63
Identities = 143/352 (40%), Positives = 195/352 (54%), Gaps = 6/352 (1%)
Frame = +1
Query: 256 QGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKS 315
+GR AG GMIL N +G+ L+A+S++L + G IG + S
Sbjct: 13 KGRVVKEAGGIGMILANTAA-SGEELVADSHLLPAVAV--------GRIIGDQIRKYVSS 165
Query: 316 VIKIR--MSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASV 373
+ +S T +P+PV+A+FSSRGPN + ILKPDV PGVNILA +S
Sbjct: 166 DLNPTTVLSFGGTVLNVRPSPVVAAFSSRGPNMITKEILKPDVIGPGVNILAGWSEAVGP 345
Query: 374 SNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTN 433
S L D R+ FNI GTSMSCPH++G A L+K HP WSP+AIKSA+MTTA DN+
Sbjct: 346 SGLAEDTRKT-KFNIMSGTSMSCPHISGLAALLKAAHPTWSPSAIKSALMTTAYNHDNSK 522
Query: 434 KLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLN 493
+RDA D + + P A+G+GH+ P A+ PGLVYD S DY+ FLC+ Y+ I ++
Sbjct: 523 SPLRDAADGSFSTPLAHGAGHVNPQKALSPGLVYDASTKDYITFLCSLNYNSEQIQLIVK 702
Query: 494 PNMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPP-STYFAKVQLP-GYNIV 551
+ LNYPS ++ V TRIVTNVG S Y V +P I
Sbjct: 703 RPSVNCTKKFANPGQLNYPSFSVVFSSKRVVRYTRIVTNVGEAGSVYNVVVDVPSSVGIT 882
Query: 552 VVPDSLTFKKNGEKKKFQV--IVQARSVTPRGRYQFGELQWTNGKHIVRSPV 601
V P L F+K GE+K++ V + + + + R FG + W+N +H VRSP+
Sbjct: 883 VKPSRLVFEKVGERKRYTVTFVSKKGADASKVRSGFGSILWSNAQHQVRSPI 1038
>TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase (EC
3.4.21.-) 2 - tomato, partial (36%)
Length = 1062
Score = 186 bits (472), Expect(2) = 2e-54
Identities = 96/187 (51%), Positives = 126/187 (67%), Gaps = 1/187 (0%)
Frame = +2
Query: 331 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQ 390
+P+P++A+FSSRGP+ + ILKPD+ APGVNILAA+S S+L D+RR FNI
Sbjct: 44 RPSPIVAAFSSRGPSLLTLEILKPDIVAPGVNILAAWSGLTGPSSLPIDHRR-VKFNILS 220
Query: 391 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAY 450
GTSMSCPHV+G A +IK HP WSPAAIKSAIMTTA + DNT K +RDA + P+ +
Sbjct: 221 GTSMSCPHVSGIAAMIKAKHPEWSPAAIKSAIMTTAYVHDNTIKPLRDASSAEFSTPYDH 400
Query: 451 GSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCS-GIHSINDL 509
G+GHI P A+DPGL+YD+ DY FLC S + + + N C + S +DL
Sbjct: 401 GAGHINPRKALDPGLLYDIEPQDYFEFLCTKKLSPSEL-VVFSKNSNRNCKHTLASASDL 577
Query: 510 NYPSITL 516
NYP+I++
Sbjct: 578 NYPAISV 598
Score = 45.4 bits (106), Expect(2) = 2e-54
Identities = 26/81 (32%), Positives = 43/81 (52%), Gaps = 2/81 (2%)
Frame = +1
Query: 525 NVTRIVTNVGPPSTYFAKVQLP--GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGR 582
N+ R VTNVGP + + + P G + V PD+L F + +K +++ + S +
Sbjct: 631 NIHRTVTNVGPAVSKYHVIGTPFKGAVVKVEPDTLNFTRKYQKLSYKISFKVTS--RQSE 804
Query: 583 YQFGELQWTNGKHIVRSPVTV 603
+FG L W + H VRSP+ +
Sbjct: 805 PEFGGLVWKDRLHKVRSPIVI 867
>TC79133 similar to GP|14150446|gb|AAK53065.1 subtilisin-type protease
precursor {Glycine max}, partial (42%)
Length = 1101
Score = 204 bits (519), Expect = 8e-53
Identities = 120/279 (43%), Positives = 170/279 (60%), Gaps = 9/279 (3%)
Frame = +1
Query: 331 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQ 390
KPAP++A FSSRGP+ + ILKPD+ APGV ILAA+ + V ++ P+ ++
Sbjct: 145 KPAPMVAIFSSRGPSALSKNILKPDIAAPGVTILAAW--IGNDDENVPKGKKPLPYKLET 318
Query: 391 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAY 450
GTSMSCPHV+G AG IK+ +P WS +AI+SAIMT+AT +N I + ++A P+ Y
Sbjct: 319 GTSMSCPHVSGLAGSIKSRNPTWSASAIRSAIMTSATQINNMKAPITTDLG-SVATPYDY 495
Query: 451 GSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLN--PNMTFTC---SGIHS 505
G+G I + PGLVY+ S +DYLN+LC GY+ I + P+ TF C S
Sbjct: 496 GAGDITTIESFQPGLVYETSTIDYLNYLCYIGYNTTTIKVISKTVPD-TFNCPKESTPDH 672
Query: 506 INDLNYPSITLPNL-GLNAVNVTRIVTNVGPPS--TYFAKVQLP-GYNIVVVPDSLTFKK 561
I+++NYPSI + N G VNV+R VTNVG Y A V P G + ++P+ L F K
Sbjct: 673 ISNINYPSIAISNFTGKETVNVSRTVTNVGEEDEVAYSAIVNAPSGVKVQLIPEKLQFTK 852
Query: 562 NGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSP 600
+ +K+ +Q I + ++T FG + W+NGK+ VRSP
Sbjct: 853 SNKKQSYQAIF-STTLTSLKEDLFGSITWSNGKYSVRSP 966
>TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase (EC
3.4.21.-) p69f - tomato, partial (27%)
Length = 1097
Score = 192 bits (487), Expect = 4e-49
Identities = 130/306 (42%), Positives = 175/306 (56%), Gaps = 14/306 (4%)
Frame = +2
Query: 312 TIKSVIKIRMSQPKTSYRRKP------APVMASFSSRGPNQVQPYILKPDVTAPGVNILA 365
TIKS IK + T + AP + FSSRGP+Q P ILKPD+ PGVNILA
Sbjct: 56 TIKSYIKSTYNPTATLIFKGTIIGDSLAPSVVYFSSRGPSQESPGILKPDIIGPGVNILA 235
Query: 366 AYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 425
A+++ SV N + F+I GTSMSCPH++G A LIK+ HP+WSPAAIKSAIMTT
Sbjct: 236 AWAV--SVDNKIP------AFDIVSGTSMSCPHLSGIAALIKSSHPDWSPAAIKSAIMTT 391
Query: 426 ATIRDNTNKLIRDAIDKTL--ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGY 483
A + N +D+ L A+ FA G+GH+ P A DPGLVYD+ DY+ +LC GY
Sbjct: 392 A---NTLNLGGIPILDQRLLPADIFATGAGHVNPFKANDPGLVYDIEPEDYVPYLCGLGY 562
Query: 484 SQRLISTLLNPNMTFTCSGIHSIND--LNYPSITLPNLGLNAVNVTRIVTNVG-PPSTYF 540
S + I ++ CS + SI + LNYPS ++ LG ++ TR +TNVG STY
Sbjct: 563 SDKEIEVIV--QWKVKCSNVKSIPEAQLNYPSFSI-LLGSDSQYYTRTLTNVGFANSTYK 733
Query: 541 AKVQLP-GYNIVVVPDSLTFKKNGEKKKFQV--IVQARSVTPRGRYQFGELQWTNGKHIV 597
++++P + V P +TF + EK F V I Q + + G L W + KH V
Sbjct: 734 VELEVPLALGMSVNPSEITFTEVNEKVSFSVEFIPQIKENRRNQTFGQGSLTWVSDKHAV 913
Query: 598 RSPVTV 603
R P++V
Sbjct: 914 RVPISV 931
>TC81650 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (19%)
Length = 972
Score = 191 bits (486), Expect = 5e-49
Identities = 114/261 (43%), Positives = 151/261 (57%), Gaps = 10/261 (3%)
Frame = +2
Query: 354 PDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNW 413
PDV APGVNILA ++ ++L D RR FNI GTSMSCPHV+G A L++ +P W
Sbjct: 2 PDVIAPGVNILAGWTGKVGPTDLEIDPRR-VEFNIISGTSMSCPHVSGIAALLRKAYPEW 178
Query: 414 SPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVD 473
SPAAIKSA+MTTA DN+ I+D +NPF +G+GH+ PN A++PGLVYDL++ D
Sbjct: 179 SPAAIKSALMTTAYNVDNSGGKIKDLGTGKESNPFVHGAGHVDPNKALNPGLVYDLNIND 358
Query: 474 YLNFLCAAGYSQRLISTLL-NPNMTFTCSG---IHSINDLNYPSITL---PNLGLNAVNV 526
YL FLC+ GY + I P C S DLNYPS ++ N GL V
Sbjct: 359 YLAFLCSIGYDAKEIQIFTREPTSYNVCENERKFTSPGDLNYPSFSVVFGANNGL--VKY 532
Query: 527 TRIVTNVGP--PSTYFAKVQLP-GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRY 583
R++TNVG + Y KV P G ++ V P L F + + F+V + G
Sbjct: 533 KRVLTNVGDSVDAVYTVKVNAPFGVDVSVSPSKLVFSSENKTQAFEVTF--TRIGYGGSQ 706
Query: 584 QFGELQWTNGKHIVRSPVTVQ 604
FG L+W++G HIVRSP+ +
Sbjct: 707 SFGSLEWSDGSHIVRSPIAAR 769
>TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycopersicon
esculentum}, partial (37%)
Length = 1230
Score = 191 bits (486), Expect = 5e-49
Identities = 147/419 (35%), Positives = 211/419 (50%), Gaps = 17/419 (4%)
Frame = +3
Query: 202 NVTNQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREAL 261
N ++ FC P + V GKIV C G +G V++G+
Sbjct: 21 NTSDDSIAFCGPIAMKKVDVKGKIVVCE-------------QGGFVG----RVAKGQAVK 149
Query: 262 SAGAKGMILRNQP--KFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKI 319
AG MIL N FN +A+ +VL ++ +S G++ D I S
Sbjct: 150 DAGGAAMILLNSEGEDFNP---IADVHVLPAVHV--------SYSAGLNIQDYINST--- 287
Query: 320 RMSQPKTSYRRKP-------APVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFAS 372
S P + K AP +ASFSSRGP++ P ILKPD+ PG+NILA + + S
Sbjct: 288 --STPMATILFKGTVIGNPNAPQVASFSSRGPSKASPGILKPDILGPGLNILAGWPI--S 455
Query: 373 VSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNT 432
+ N + FNI GTSMSCPH++G A L+K HP+WSPAAIKSAIMTTA ++
Sbjct: 456 LDNSTSS------FNIIAGTSMSCPHLSGIAALLKNSHPDWSPAAIKSAIMTTA---NHV 608
Query: 433 NKLIRDAIDKTL--ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLIST 490
N + +D+ L A+ FA G+GH+ P+ A DPGLVYD+ DY+ +LC Y+ +
Sbjct: 609 NLHGKPILDQRLLPADVFATGAGHVNPSKANDPGLVYDIETNDYVPYLCGLNYTDIQVGI 788
Query: 491 LLNPNMTFTCSGIHSI--NDLNYPSITLPNLGLNAVNVTRIVTNVGP-PSTYFAKVQLP- 546
+L + CS + SI LNYPSI++ LG + +R +TNVGP +TY + +P
Sbjct: 789 ILQQKV--KCSDVKSIPQAQLNYPSISI-RLGNTSQFYSRTLTNVGPVNTTYNVVIDVPV 959
Query: 547 GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGR--YQFGELQWTNGKHIVRSPVTV 603
+ V P +TF + +K + V RG G ++W + K+ V P+ V
Sbjct: 960 AVRMSVRPSQITFTEVKQKVTYWVDFIPEDKENRGDNFIAQGSIKWISAKYSVSIPIAV 1136
>CB892799 weakly similar to PIR|T05768|T05 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (20%)
Length = 875
Score = 183 bits (464), Expect = 2e-46
Identities = 128/323 (39%), Positives = 180/323 (55%), Gaps = 3/323 (0%)
Frame = +2
Query: 117 DEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITIGNK- 175
D ISIG+F A + + +V+SAGN G GS TN+APW+ TVAA + DRDF+S I +GN
Sbjct: 2 DAISIGSFHAANRGLFVVSSAGNEGNL-GSATNLAPWMLTVAAGSTDRDFTSDIILGNGA 178
Query: 176 --TGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSKVSGKIVECVGEKI 233
TG SL + N S ++ + +A T + +C +L+ +K GK++ C
Sbjct: 179 KITGESLSL-FEMNASTRIISASEAFAGYFTPYQSSYCLESSLNKTKTKGKVLVCRH--- 346
Query: 234 TIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTINY 293
++ ++E + V++ + AG GMIL ++ + +A V+ +
Sbjct: 347 -VERSTE-----------SKVAKSKIVKEAGGVGMILIDETDQD----VAIPFVIPSAIV 478
Query: 294 YDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILK 353
K +G I T K + KI + KT + AP +A+FSSRGPN + P ILK
Sbjct: 479 GKK----KGQKILSYLKTTRKPMSKILRA--KTVIGAQSAPRVAAFSSRGPNALNPEILK 640
Query: 354 PDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNW 413
PD+TAPG+NILAA+S A G FNI GTSM+CPHV G A L+K +HP+W
Sbjct: 641 PDITAPGLNILAAWSPVA-----------GNMFNILSGTSMACPHVTGIATLVKAVHPSW 787
Query: 414 SPAAIKSAIMTTATIRDNTNKLI 436
SP+AIKSAIMTTATI D +K I
Sbjct: 788 SPSAIKSAIMTTATILDKRHKPI 856
>BG580583 weakly similar to GP|9757901|dbj serine protease-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 592
Score = 162 bits (410), Expect(2) = 2e-46
Identities = 82/114 (71%), Positives = 93/114 (80%)
Frame = +3
Query: 32 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 91
TAGGNFV A+IFGIGNGTIKGGSP+SRV TYK CWS T + C+GADVL+AIDQ+
Sbjct: 261 TAGGNFVQNATIFGIGNGTIKGGSPRSRVATYKACWSLT----DVVDCFGADVLAAIDQS 428
Query: 92 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPG 145
I DG D+ISVS GG+ ++N E IFTDEISIGAF A A+NILLVASAGN GPTPG
Sbjct: 429 IYDGADLISVSAGGKPNTNPEVIFTDEISIGAFHALARNILLVASAGNEGPTPG 590
Score = 42.0 bits (97), Expect(2) = 2e-46
Identities = 25/41 (60%), Positives = 27/41 (64%), Gaps = 9/41 (21%)
Frame = +1
Query: 1 VFGPNRRVLTTEE*VQFQQNGVGE---------VQRKFHVT 32
VFG NRRVLTTEE*VQF +GVG + RKFHVT
Sbjct: 22 VFGRNRRVLTTEE*VQFH*DGVGVTFVNLTNSILLRKFHVT 144
>TC83642 similar to PIR|S52769|S52769 subtilisin-like proteinase ag12 (EC
3.4.21.-) - alder, partial (30%)
Length = 1177
Score = 129 bits (324), Expect(2) = 4e-43
Identities = 77/166 (46%), Positives = 99/166 (59%), Gaps = 5/166 (3%)
Frame = +3
Query: 386 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRD-AIDKTL 444
+N GTSMSCPHV+G A L+K HP WS AAI+SA++TTA DNT IRD
Sbjct: 624 YNFMSGTSMSCPHVSGVAALLKAAHPQWSAAAIRSALITTANPLDNTQNPIRDNGYPSQH 803
Query: 445 ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIH 504
A+P A G+G I PN AM+PGL+YD + DY+N LC +++ I T+ N ++ C +
Sbjct: 804 ASPLAIGAGEIDPNRAMNPGLIYDATPQDYVNLLCGLKFTKNQILTITRSN-SYDCE--N 974
Query: 505 SINDLNYPSIT--LPNLGLNAVN-VTRIVTNVGP-PSTYFAKVQLP 546
DLNYPS N + V+ RIVTNVG +TY AKV P
Sbjct: 975 PSLDLNYPSFIAFYSNKTRSMVHKFKRIVTNVGDGAATYRAKVTYP 1112
Score = 64.3 bits (155), Expect(2) = 4e-43
Identities = 60/226 (26%), Positives = 99/226 (43%), Gaps = 7/226 (3%)
Frame = +2
Query: 149 NVAPWVFTVAASTIDRDFSSTITIGNKTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 208
N PW+ T AA TIDR F T+ +GN + P N +V+++ + N +
Sbjct: 2 NGIPWLLTAAAGTIDRTF-GTLVLGNGQSIIGWTLFPAN---AIVENVLLVYNNTLSS-- 163
Query: 209 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 268
C L S+++ K++ + ++ +N + N ++ EA GA +
Sbjct: 164 --CNSLNL-LSQLNKKVIILCDDSLSNRNKTS---------VFNQINVVTEANLLGA--V 301
Query: 269 ILRNQPK-------FNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRM 321
+ + P+ + ++ + S INY + + T +IK
Sbjct: 302 FVSDSPQLIDLGRIYTPSIVIKPKDAQSVINYAKSNN---------NPTSSIKF------ 436
Query: 322 SQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAY 367
+T KPAP A +SSRGP+ P+ILKPD+ APG +LAAY
Sbjct: 437 --QQTFVGTKPAPAAAYYSSRGPSHSYPWILKPDIMAPGSRVLAAY 568
>TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like serine
protease AIR3 {Arabidopsis thaliana}, partial (16%)
Length = 704
Score = 148 bits (373), Expect(2) = 3e-41
Identities = 75/97 (77%), Positives = 82/97 (84%)
Frame = +1
Query: 32 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 91
TAGGNFVPGASIF IGNGTIKGGSP++RV TYKVCWS T ++A C+GADVLSAIDQA
Sbjct: 424 TAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLT----DAASCFGADVLSAIDQA 591
Query: 92 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFA 128
I DGVDII VS GG SS+N EEIFTDE+SIGAF A A
Sbjct: 592 IDDGVDIIFVSAGGPSSTNSEEIFTDEVSIGAFHALA 702
Score = 39.3 bits (90), Expect(2) = 3e-41
Identities = 24/41 (58%), Positives = 25/41 (60%), Gaps = 9/41 (21%)
Frame = +2
Query: 1 VFGPNRRVLTTEE*VQFQQNGVG---------EVQRKFHVT 32
VFG N+ VL TEE QFQQNGVG EV RKF VT
Sbjct: 185 VFGRNQGVLATEELAQFQQNGVGVMSVKLTNSEVLRKFLVT 307
>TC88594 similar to PIR|T08978|T08978 serine proteinase homolog F6G3.50 -
Arabidopsis thaliana, partial (37%)
Length = 1049
Score = 165 bits (418), Expect = 4e-41
Identities = 115/286 (40%), Positives = 148/286 (51%), Gaps = 15/286 (5%)
Frame = +2
Query: 331 KPAPVMASFSSRGPNQ-----VQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 385
K AP +A FS+RGPN + +LKPD+ APG I A+S + N G
Sbjct: 89 KSAPQVALFSARGPNIRDFSFQEADLLKPDILAPGSLIWGAWSR----NGTDEPNYDGEG 256
Query: 386 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLI-------RD 438
F + GTSM+ PH+AG A LIK HP WSPAAIKSA++TT T D I +
Sbjct: 257 FAMVSGTSMAAPHIAGIAALIKQKHPRWSPAAIKSALLTTTTTLDRGGNPILSQQYSETE 436
Query: 439 AIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAA-GYSQRLISTLLNPNMT 497
A+ A PF YG+GH+ P A+DPGL++D DYL FLC G I N
Sbjct: 437 AMKLVKATPFDYGNGHVNPRAALDPGLIFDAGYKDYLGFLCTTPGIDVHEIKKYTNSPCN 616
Query: 498 FTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQL-PGYNIVVVPDS 556
T H N LN PSIT+ +L + +TR VTNV TY ++ P I + P +
Sbjct: 617 RTMG--HPYN-LNTPSITVSHL-VRTQTITRKVTNVAKEETYVLTARMQPAVAIEITPPA 784
Query: 557 LTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGK-HIVRSPV 601
+T + G +KF V + RSVT G Y FGE+ + H VR PV
Sbjct: 785 MTIRA-GASRKFTVTLTVRSVT--GTYSFGEVLMKGSRGHKVRIPV 913
>BI307979 weakly similar to GP|20521377|db putative subtilisin-like protease
{Oryza sativa (japonica cultivar-group)}, partial (18%)
Length = 777
Score = 164 bits (416), Expect = 7e-41
Identities = 112/294 (38%), Positives = 158/294 (53%), Gaps = 11/294 (3%)
Frame = +3
Query: 85 LSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTP 144
L AID AI DGVD+I++S+G + +E+ D I+ GA QA KNI++V SAGN GP+P
Sbjct: 3 LKAIDDAIEDGVDVINLSIGFPAPLKYED---DVIAKGALQAVRKNIVVVCSAGNAGPSP 173
Query: 145 GSVTNVAPWVFTVAASTIDRDFSSTITIGNKT---GASLFVNLPPNQSFTLVDSIDAKFA 201
S++N +PW+ TV AST+DR F + I + N T G S+ N LV + D ++A
Sbjct: 174 HSLSNPSPWIITVGASTVDRTFLAPIKLSNGTTIEGRSITPLRMGNSFCPLVLASDVEYA 353
Query: 202 NVTNQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREAL 261
+ + ++ +C TLDPSKV GKIV C+ + GR V + E
Sbjct: 354 GILSANSSYCLDNTLDPSKVKGKIVLCMRGQ----------GGR--------VKKSLEVQ 479
Query: 262 SAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRM 321
AG G+IL N + +N + + Y+ + G++ +T+K V I
Sbjct: 480 RAGGVGLILGNNKVY--------ANDVPSDPYFIP-------TTGVTYENTLKLVQYIHS 614
Query: 322 S--------QPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAY 367
S +T KPAP MA FSSRGPN + P ILKPD+TAPGV+I AA+
Sbjct: 615 SPNPMAQLLPGRTVLDTKPAPSMAIFSSRGPNIIDPNILKPDITAPGVDIFAAW 776
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,833,303
Number of Sequences: 36976
Number of extensions: 199783
Number of successful extensions: 1146
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1030
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1043
length of query: 605
length of database: 9,014,727
effective HSP length: 102
effective length of query: 503
effective length of database: 5,243,175
effective search space: 2637317025
effective search space used: 2637317025
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135318.10


