

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135313.7 - phase: 0
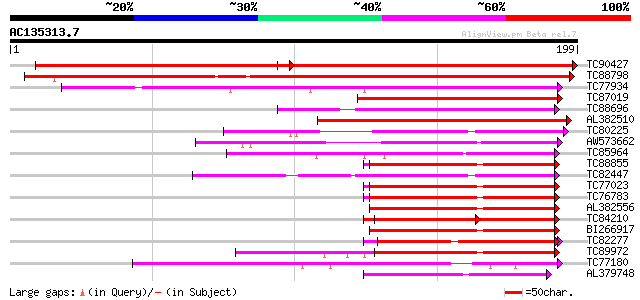
(199 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90427 weakly similar to GP|16648830|gb|AAL25605.1 AT5g39670/MI... 211 e-105
TC88798 weakly similar to GP|7939550|dbj|BAA95753.1 contains sim... 244 1e-65
TC77934 similar to GP|9965747|gb|AAG10150.1| calmodulin-like MSS... 82 1e-16
TC87019 weakly similar to GP|12003380|gb|AAG43547.1 Avr9/Cf-9 ra... 73 6e-14
TC88696 weakly similar to PIR|T10620|T10620 probable calcium-bin... 69 1e-12
AL382510 weakly similar to GP|13194672|gb| calmodulin-like prote... 69 2e-12
TC80225 similar to GP|14589311|emb|CAC43238. calcium binding pro... 62 1e-10
AW573662 weakly similar to PIR|D96689|D966 calmodulin-related pr... 60 7e-10
TC85964 similar to PIR|D84841|D84841 calmodulin-like protein [im... 57 5e-09
TC88855 PIR|A30900|MCBH calmodulin - barley, complete 54 3e-08
TC82447 similar to GP|14589311|emb|CAC43238. calcium binding pro... 54 3e-08
TC77023 calmodulin 2 [Medicago truncatula] 54 3e-08
TC76783 calmodulin 1 [Medicago truncatula] 54 3e-08
AL382556 homologue to GP|169306|gb|AA calmodulin {Phytophthora i... 52 2e-07
TC84210 homologue to PIR|T07122|T07122 calmodulin CAM5 - soybean... 51 3e-07
BI266917 PIR|S01173|MC calmodulin [validated] - fruit fly (Droso... 50 8e-07
TC82277 weakly similar to PIR|A84532|A84532 probable calmodulin-... 49 1e-06
TC89972 similar to GP|22296346|dbj|BAC10116. putative caltractin... 47 4e-06
TC77180 similar to GP|2665890|gb|AAB88537.1| calcium-dependent p... 45 2e-05
AL379748 EGAD|125673|13 calmodulin {Sus scrofa}, partial (55%) 45 2e-05
>TC90427 weakly similar to GP|16648830|gb|AAL25605.1 AT5g39670/MIJ24_140
{Arabidopsis thaliana}, partial (46%)
Length = 730
Score = 211 bits (537), Expect(2) = e-105
Identities = 102/105 (97%), Positives = 103/105 (97%)
Frame = +1
Query: 95 EKMGFFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRV 154
+K FFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRV
Sbjct: 259 KKWDFFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRV 438
Query: 155 LVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMKNHFCC 199
LVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMKNHFCC
Sbjct: 439 LVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMKNHFCC 573
Score = 186 bits (473), Expect(2) = e-105
Identities = 91/91 (100%), Positives = 91/91 (100%)
Frame = +3
Query: 10 NSISNSTSPLFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCS 69
NSISNSTSPLFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCS
Sbjct: 3 NSISNSTSPLFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCS 182
Query: 70 SQENESNIGRDNGDMIERDEVKMVMEKMGFF 100
SQENESNIGRDNGDMIERDEVKMVMEKMGFF
Sbjct: 183 SQENESNIGRDNGDMIERDEVKMVMEKMGFF 275
>TC88798 weakly similar to GP|7939550|dbj|BAA95753.1 contains similarity to
calmodulin~gene_id:K5K13.13 {Arabidopsis thaliana},
partial (45%)
Length = 804
Score = 244 bits (624), Expect = 1e-65
Identities = 122/195 (62%), Positives = 152/195 (77%), Gaps = 2/195 (1%)
Frame = +1
Query: 6 ISQSNSISN--STSPLFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKV 63
I++ N++ N S S LFGLID F+Y FF +IH FFSS WFFL CQ+ SG EV+ +K++
Sbjct: 172 IAEENTLQNDESNSTLFGLIDFFMYSMFFIRIHKFFSSFWFFLFCQLRSGDLEVKGKKQL 351
Query: 64 SESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEE 123
SE++ + +SN+ ++G+ I+R+EV+ VM MG FCSSES+ELEEKY SKE E+F+E
Sbjct: 352 SETELNVSR-DSNVSGESGE-IKRNEVETVMANMGLFCSSESDELEEKYSSKEFSELFDE 525
Query: 124 NEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGR 183
NEPSLEE+K AFDVFDENKDGFIDA EL+RV+ ILG K+GSE ENCQKMI FD NQDGR
Sbjct: 526 NEPSLEEVKMAFDVFDENKDGFIDAMELKRVMCILGFKEGSEVENCQKMIKNFDANQDGR 705
Query: 184 IDFIEFVNIMKNHFC 198
IDFIEFV IM+N C
Sbjct: 706 IDFIEFVKIMENRLC 750
>TC77934 similar to GP|9965747|gb|AAG10150.1| calmodulin-like MSS3
{Arabidopsis thaliana}, partial (80%)
Length = 1325
Score = 82.0 bits (201), Expect = 1e-16
Identities = 59/195 (30%), Positives = 95/195 (48%), Gaps = 19/195 (9%)
Frame = +1
Query: 19 LFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCSSQENESN-- 76
L+ +++ FL K+ FF WF Q + S + V + NE
Sbjct: 127 LYNVVNSFLISLVPKKLITFFPHSWF--THQTLTTPSSTSKRGLVFTKTITMDPNELKRV 300
Query: 77 ---IGRDNGDMIERDEVKMVMEKMGFFCSSE--SEELEEKYGSKELCEVFEE-------- 123
R++ I + E+ +E +G F + S+ +E+ +++ C EE
Sbjct: 301 FQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESI 480
Query: 124 ----NEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDEN 179
+E E++++AF+VFD+N DGFI EL+ VLV LGLKQG E+C+KMI D +
Sbjct: 481 MSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVD 660
Query: 180 QDGRIDFIEFVNIMK 194
+G +D+ EF +MK
Sbjct: 661 GNGLVDYKEFKQMMK 705
>TC87019 weakly similar to GP|12003380|gb|AAG43547.1 Avr9/Cf-9 rapidly
elicited protein 31 {Nicotiana tabacum}, partial (76%)
Length = 967
Score = 73.2 bits (178), Expect = 6e-14
Identities = 37/72 (51%), Positives = 50/72 (69%)
Frame = +3
Query: 123 ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDG 182
+ E E+L +AF VFDE+ DG+I AKELQ VL LGL +G+ +N Q+MI D N DG
Sbjct: 498 DEETQNEDLWEAFKVFDEDGDGYISAKELQVVLGKLGLVEGNLIDNVQRMILSVDTNHDG 677
Query: 183 RIDFIEFVNIMK 194
R+DF EF ++M+
Sbjct: 678 RVDFHEFKDMMR 713
Score = 37.4 bits (85), Expect = 0.004
Identities = 19/62 (30%), Positives = 37/62 (59%)
Frame = +3
Query: 131 LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFV 190
L++ FD+FD+N D I +E+ + L +LGL+ +EF+ MI + + + + + +FV
Sbjct: 273 LRRIFDMFDKNGDSMITVEEISQALNLLGLE--AEFKEVDSMIKSYIKPGNVGLTYEDFV 446
Query: 191 NI 192
+
Sbjct: 447 GL 452
>TC88696 weakly similar to PIR|T10620|T10620 probable calcium-binding
protein F21C20.130 - Arabidopsis thaliana, partial (17%)
Length = 648
Score = 68.9 bits (167), Expect = 1e-12
Identities = 40/99 (40%), Positives = 57/99 (57%)
Frame = +1
Query: 95 EKMGFFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRV 154
E + F+ S E+ EE G E +EN+ +L + F VFD + DGFI ++EL+ V
Sbjct: 226 EFLFFYNSISKEKNEENRGGDE-----DENDELERDLVKTFKVFDLDGDGFITSQELECV 390
Query: 155 LVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIM 193
L LG S ++C+ MI +D N DGR+DF EF N+M
Sbjct: 391 LKRLGFLDESSGKDCRSMIRFYDTNLDGRLDFQEFKNMM 507
>AL382510 weakly similar to GP|13194672|gb| calmodulin-like protein
{Pennisetum ciliare}, partial (24%)
Length = 399
Score = 68.6 bits (166), Expect = 2e-12
Identities = 36/89 (40%), Positives = 55/89 (61%)
Frame = +1
Query: 109 EEKYGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFEN 168
EE+ +E+ E+ E L +AF +FDE+ DG+IDA EL+RVL LGL +G +
Sbjct: 31 EEEVVVEEVLGELEDMSKKSELLLEAFKIFDEDGDGYIDAMELKRVLDCLGLDKGWDMNT 210
Query: 169 CQKMITIFDENQDGRIDFIEFVNIMKNHF 197
++M+ + D N DG++DF EF +M +F
Sbjct: 211 IERMVKVVDLNFDGKVDFGEFELMMG*YF 297
>TC80225 similar to GP|14589311|emb|CAC43238. calcium binding protein
{Sesbania rostrata}, partial (87%)
Length = 704
Score = 62.0 bits (149), Expect = 1e-10
Identities = 44/134 (32%), Positives = 66/134 (48%), Gaps = 13/134 (9%)
Frame = +3
Query: 76 NIGRDNGDMIERDEVKMVMEKM-----GF--------FCSSESEELEEKYGSKELCEVFE 122
NI R G + +DE++ VME + GF FC S S + +
Sbjct: 153 NILRSLGSTVPKDELQRVMEDLDTDRDGFINLAEFAAFCRSGSADGD------------- 293
Query: 123 ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDG 182
+ EL++AFD++D++K+G I A EL +VL LG+K E C MI D + DG
Sbjct: 294 -----VSELREAFDLYDKDKNGLISATELCQVLNTLGMK--CSVEECHTMIKSVDSDGDG 452
Query: 183 RIDFIEFVNIMKNH 196
++F EF +M N+
Sbjct: 453 NVNFEEFKKMMNNN 494
>AW573662 weakly similar to PIR|D96689|D966 calmodulin-related protein
72976-72503 [imported] - Arabidopsis thaliana, partial
(29%)
Length = 615
Score = 59.7 bits (143), Expect = 7e-10
Identities = 45/133 (33%), Positives = 69/133 (51%), Gaps = 4/133 (3%)
Frame = +1
Query: 66 SKCSSQENESNIGRD---NGD-MIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVF 121
+KC++ + ++ R NGD ++ DE VM++ G F + + +E
Sbjct: 205 NKCTASKEAESMIRVLDFNGDGFVDIDEFMFVMKEDGNFGKGKEHDHDEY---------- 354
Query: 122 EENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQD 181
L AF VFD +K+G I KEL+RVLV LG + S +C++MI D+N D
Sbjct: 355 ---------LMDAFLVFDIDKNGLISPKELRRVLVNLGCENCS-LRDCKRMIKGVDKNGD 504
Query: 182 GRIDFIEFVNIMK 194
G +DF EF ++MK
Sbjct: 505 GFVDFEEFRSMMK 543
>TC85964 similar to PIR|D84841|D84841 calmodulin-like protein [imported] -
Arabidopsis thaliana, partial (69%)
Length = 825
Score = 57.0 bits (136), Expect = 5e-09
Identities = 43/133 (32%), Positives = 69/133 (51%), Gaps = 16/133 (12%)
Frame = +1
Query: 77 IGRDNGDMIERDEVKMVMEKMGFFCSSESE---ELEEKYGSKELCEVFEE---------- 123
I RDN ++ R+E++ V+ ++G + E L E + C E
Sbjct: 304 IDRDNDGVVSREELEAVLTRLGARPPTPEEIALMLSEVDSDGKGCISVETIMNRVGSGSS 483
Query: 124 --NEPSLEE-LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQ 180
++P+ EE L++AF+VFD ++DG I A+EL RV +G + E C++MI D+N
Sbjct: 484 SGSDPNPEEELREAFEVFDTDRDGRISAEELLRVFRAIG-DERCTLEECKRMIAGVDKNG 660
Query: 181 DGRIDFIEFVNIM 193
DG + F EF +M
Sbjct: 661 DGFVCFQEFSLMM 699
>TC88855 PIR|A30900|MCBH calmodulin - barley, complete
Length = 889
Score = 54.3 bits (129), Expect = 3e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 319 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 492
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 493 EEFVKVM 513
Score = 48.5 bits (114), Expect = 2e-06
Identities = 28/69 (40%), Positives = 39/69 (55%)
Frame = +1
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 94 DDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 267
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 268 DFPEFLNLM 294
>TC82447 similar to GP|14589311|emb|CAC43238. calcium binding protein
{Sesbania rostrata}, partial (79%)
Length = 660
Score = 54.3 bits (129), Expect = 3e-08
Identities = 37/129 (28%), Positives = 65/129 (49%)
Frame = +2
Query: 65 ESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEEN 124
+ K S E E+ + D+ +++E++ VME++ S++ + C +
Sbjct: 218 DGKISVNELETVLRTLRSDVPQQEELRRVMEEL----STDRDGFINLSEFAAFCRS-DTT 382
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ L+ AFD++D++K+G I +EL L LG+K E CQ MI D + DG +
Sbjct: 383 DGGDSALRDAFDLYDKDKNGLISTEELHLALDRLGMK--CSVEECQDMIKSVDSDGDGSV 556
Query: 185 DFIEFVNIM 193
+F EF +M
Sbjct: 557 NFDEFKKMM 583
>TC77023 calmodulin 2 [Medicago truncatula]
Length = 999
Score = 54.3 bits (129), Expect = 3e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 328 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 501
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 502 EEFVKVM 522
Score = 48.9 bits (115), Expect = 1e-06
Identities = 28/69 (40%), Positives = 39/69 (55%)
Frame = +1
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 103 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 276
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 277 DFPEFLNLM 303
>TC76783 calmodulin 1 [Medicago truncatula]
Length = 957
Score = 54.3 bits (129), Expect = 3e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +3
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 363 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 536
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 537 EEFVKVM 557
Score = 48.9 bits (115), Expect = 1e-06
Identities = 28/69 (40%), Positives = 39/69 (55%)
Frame = +3
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 138 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 311
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 312 DFPEFLNLM 338
>AL382556 homologue to GP|169306|gb|AA calmodulin {Phytophthora infestans},
partial (49%)
Length = 476
Score = 52.0 bits (123), Expect = 2e-07
Identities = 30/67 (44%), Positives = 43/67 (63%)
Frame = +2
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EE+K+AF VFD++ +GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 20 SEEEIKEAFKVFDKDGNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 193
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 194 EEFVKMM 214
>TC84210 homologue to PIR|T07122|T07122 calmodulin CAM5 - soybean, partial
(96%)
Length = 627
Score = 51.2 bits (121), Expect = 3e-07
Identities = 28/69 (40%), Positives = 42/69 (60%)
Frame = +1
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
E + E+K+AF +FD++ DG I +EL VI L Q E Q+MI D + +G I
Sbjct: 1 EDQIVEIKEAFCLFDKDGDGCITVEEL--ATVIRSLDQNPTEEELQEMINEVDADGNGTI 174
Query: 185 DFIEFVNIM 193
+F+EF+N+M
Sbjct: 175 EFVEFLNLM 201
Score = 40.4 bits (93), Expect = 5e-04
Identities = 18/37 (48%), Positives = 28/37 (75%)
Frame = +1
Query: 129 EELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSE 165
E+LK+AF VFD++++G+I A EL+ V++ LG K E
Sbjct: 232 EDLKEAFKVFDKDQNGYISASELRHVMINLGEKLTDE 342
>BI266917 PIR|S01173|MC calmodulin [validated] - fruit fly (Drosophila
melanogaster), complete
Length = 576
Score = 49.7 bits (117), Expect = 8e-07
Identities = 28/67 (41%), Positives = 43/67 (63%)
Frame = +2
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EE+++AF VFD++ +GFI A EL+ V+ LG K E +MI D + DG++++
Sbjct: 254 SEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADIDGDGQVNY 427
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 428 EEFVTMM 448
>TC82277 weakly similar to PIR|A84532|A84532 probable calmodulin-like
protein [imported] - Arabidopsis thaliana, partial (91%)
Length = 780
Score = 49.3 bits (116), Expect = 1e-06
Identities = 26/70 (37%), Positives = 39/70 (55%)
Frame = +3
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+PSL+E+K FD FD NKDG I +E + L LG+++ + + D + DG I
Sbjct: 159 QPSLDEMKMVFDKFDSNKDGKISQQEYKATLKSLGMEK--SVNEVPNIFRVVDLDGDGFI 332
Query: 185 DFIEFVNIMK 194
+F EF+ K
Sbjct: 333 NFEEFMEAQK 362
Score = 47.0 bits (110), Expect = 5e-06
Identities = 23/64 (35%), Positives = 40/64 (61%)
Frame = +3
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
+++ AF FD+N DG I A+E++ +L L++ E+C++M+ D + DG +D EF
Sbjct: 387 DIQTAFRTFDKNGDGKISAEEIKEML--WKLEERCSLEDCRRMVRAVDTDGDGMVDMNEF 560
Query: 190 VNIM 193
V +M
Sbjct: 561 VAMM 572
>TC89972 similar to GP|22296346|dbj|BAC10116. putative caltractin {Oryza
sativa (japonica cultivar-group)}, partial (33%)
Length = 801
Score = 47.4 bits (111), Expect = 4e-06
Identities = 25/65 (38%), Positives = 40/65 (61%)
Frame = +1
Query: 129 EELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIE 188
+E+K+AF++FD + G IDAKEL + LG + E ++MI D++ G ID+ E
Sbjct: 133 QEIKEAFELFDTDGSGTIDAKELNVAMRALGFEMTE--EQIEQMIADVDKDGSGAIDYDE 306
Query: 189 FVNIM 193
F ++M
Sbjct: 307 FEHMM 321
Score = 45.8 bits (107), Expect = 1e-05
Identities = 36/126 (28%), Positives = 60/126 (47%), Gaps = 12/126 (9%)
Frame = +1
Query: 80 DNGDMIERDEVKMVMEKMGFFCSSESEELE----EKYGSKEL-CEVFEE-------NEPS 127
D I+ E+ + M +GF + E E +K GS + + FE +
Sbjct: 169 DGSGTIDAKELNVAMRALGFEMTEEQIEQMIADVDKDGSGAIDYDEFEHMMTAKIGERDT 348
Query: 128 LEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFI 187
EEL +AF + D++K+G I A +++R+ LG Q Q+M+ D+N D +D
Sbjct: 349 KEELMKAFHIIDKDKNGKISASDIKRIAKELG--QNFTDREIQEMVDEADQNNDREVDPE 522
Query: 188 EFVNIM 193
EF+ +M
Sbjct: 523 EFIMMM 540
>TC77180 similar to GP|2665890|gb|AAB88537.1| calcium-dependent protein
kinase {Fragaria x ananassa}, partial (41%)
Length = 681
Score = 45.1 bits (105), Expect = 2e-05
Identities = 43/167 (25%), Positives = 74/167 (43%), Gaps = 16/167 (9%)
Frame = +1
Query: 44 FFLLCQIHSGSSEVREEKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFC-S 102
F ++ ++ + V E E +E + + N I DE++ + K+G
Sbjct: 172 FSVMNKLKKRALRVIAEHLSVEEAAGLKEGFNLMDTTNRGKINIDELRTGLHKLGHQVPD 351
Query: 103 SESEELEEK----------YGSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQ 152
S+ + L E YG V + E L +AFD FD+N+ G+I+ +EL+
Sbjct: 352 SDLQILMEAGDIDRDGYLDYGEYVAISVHLRKMGNDEHLHKAFDFFDQNQTGYIEIEELR 531
Query: 153 RVLVILGLKQGSEFE-NCQKMITIF----DENQDGRIDFIEFVNIMK 194
L E E N +++I+ D ++DG+I + EF N+MK
Sbjct: 532 NAL-------SDEIETNSEEVISAIMHDVDTDKDGKISYEEFANMMK 651
>AL379748 EGAD|125673|13 calmodulin {Sus scrofa}, partial (55%)
Length = 262
Score = 44.7 bits (104), Expect = 2e-05
Identities = 27/66 (40%), Positives = 36/66 (53%)
Frame = +1
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
E + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 70 EEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 243
Query: 185 DFIEFV 190
DF EF+
Sbjct: 244 DFPEFL 261
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,401,492
Number of Sequences: 36976
Number of extensions: 88783
Number of successful extensions: 886
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 837
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 860
length of query: 199
length of database: 9,014,727
effective HSP length: 91
effective length of query: 108
effective length of database: 5,649,911
effective search space: 610190388
effective search space used: 610190388
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135313.7


