

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135311.3 - phase: 1 /pseudo
(84 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
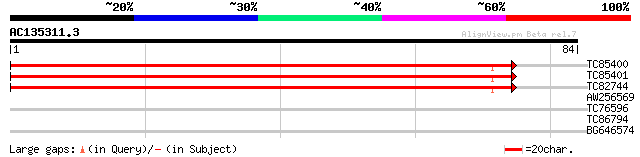
Score E
Sequences producing significant alignments: (bits) Value
TC85400 sucrose synthase 132 3e-32
TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase ... 110 1e-25
TC82744 homologue to EGAD|142043|151512 second sucrose synthase ... 98 5e-22
AW256569 similar to GP|10177426|dbj GATA-binding transcription f... 26 3.1
TC76596 homologue to SP|P33278|SUI1_ORYSA Protein translation fa... 25 5.3
TC86794 similar to PIR|T06499|T06499 Rieske [2Fe-2S] iron-sulfur... 25 5.3
BG646574 weakly similar to GP|6520167|dbj| ZW18 {Arabidopsis tha... 24 9.1
>TC85400 sucrose synthase
Length = 3133
Score = 132 bits (331), Expect = 3e-32
Identities = 69/80 (86%), Positives = 70/80 (87%), Gaps = 5/80 (6%)
Frame = +2
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IVLPPWVALAVR PGI EYLRVNVHALVVENLQPAEFLKFKEELVDGSAN N+VLELDF
Sbjct: 548 IVLPPWVALAVRPRPGIWEYLRVNVHALVVENLQPAEFLKFKEELVDGSANGNFVLELDF 727
Query: 61 EPFTASFPRP-----IGGGV 75
EPFTASFPRP IG GV
Sbjct: 728 EPFTASFPRPTLNKSIGNGV 787
>TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase isoform 3
{Pisum sativum}, complete
Length = 2846
Score = 110 bits (275), Expect = 1e-25
Identities = 56/80 (70%), Positives = 67/80 (83%), Gaps = 5/80 (6%)
Frame = +2
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
+VLPP+VALAVR PG+ EYLRV+VH+LVV+ L+ AE+LKFKEELV+GS+N N+VLELDF
Sbjct: 368 VVLPPFVALAVRPRPGVWEYLRVDVHSLVVDELRAAEYLKFKEELVEGSSNGNFVLELDF 547
Query: 61 EPFTASFPRP-----IGGGV 75
EPF ASFPRP IG GV
Sbjct: 548 EPFNASFPRPTLNKSIGNGV 607
>TC82744 homologue to EGAD|142043|151512 second sucrose synthase {Pisum
sativum}, partial (19%)
Length = 676
Score = 98.2 bits (243), Expect = 5e-22
Identities = 52/80 (65%), Positives = 59/80 (73%), Gaps = 5/80 (6%)
Frame = +3
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IVLPP+VA+AVR PG+ EY+RVNV L VE L +E+L FKEELVDG N N+VLELD
Sbjct: 432 IVLPPFVAIAVRPRPGVWEYVRVNVFELNVEQLSISEYLSFKEELVDGKINENFVLELDL 611
Query: 61 EPFTASFPRP-----IGGGV 75
EPF ASFPRP IG GV
Sbjct: 612 EPFNASFPRPTRSSSIGNGV 671
>AW256569 similar to GP|10177426|dbj GATA-binding transcription factor-like
protein {Arabidopsis thaliana}, partial (20%)
Length = 613
Score = 25.8 bits (55), Expect = 3.1
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +2
Query: 62 PFTASFPRPIGG 73
PF+ASFP+P GG
Sbjct: 305 PFSASFPKPYGG 340
>TC76596 homologue to SP|P33278|SUI1_ORYSA Protein translation factor SUI1
homolog (GOS2 protein). [Rice] {Oryza sativa}, partial
(74%)
Length = 1573
Score = 25.0 bits (53), Expect = 5.3
Identities = 12/37 (32%), Positives = 20/37 (53%)
Frame = +2
Query: 13 LMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGS 49
+ P E++R + + EN++ E LK EEL+ S
Sbjct: 1121 ISPSEYEHMRTAISKMSAENMELKERLKTNEELIRAS 1231
>TC86794 similar to PIR|T06499|T06499 Rieske [2Fe-2S] iron-sulfur protein
tic55 - garden pea, partial (11%)
Length = 812
Score = 25.0 bits (53), Expect = 5.3
Identities = 12/37 (32%), Positives = 20/37 (53%)
Frame = -2
Query: 13 LMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGS 49
+ P E++R + + EN++ E LK EEL+ S
Sbjct: 382 ISPSEYEHMRTAISKMSAENMELKERLKTNEELIRAS 272
>BG646574 weakly similar to GP|6520167|dbj| ZW18 {Arabidopsis thaliana},
partial (10%)
Length = 754
Score = 24.3 bits (51), Expect = 9.1
Identities = 9/35 (25%), Positives = 20/35 (56%)
Frame = +3
Query: 30 VENLQPAEFLKFKEELVDGSANVNYVLELDFEPFT 64
V+++ + L+ + D + N+NY+ E +PF+
Sbjct: 324 VQSIDGEQSLQLPDREEDATHNINYLTEESVQPFS 428
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.148 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,042,177
Number of Sequences: 36976
Number of extensions: 17537
Number of successful extensions: 89
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 89
length of query: 84
length of database: 9,014,727
effective HSP length: 60
effective length of query: 24
effective length of database: 6,796,167
effective search space: 163108008
effective search space used: 163108008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC135311.3


