

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135229.3 - phase: 0
(781 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
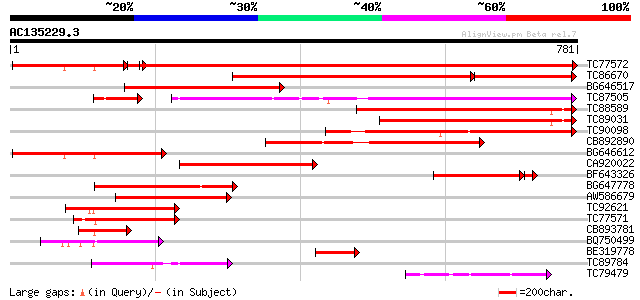
Score E
Sequences producing significant alignments: (bits) Value
TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 1108 0.0
TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 467 0.0
BG646517 405 e-113
TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein... 327 8e-90
TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 308 5e-84
TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 303 2e-82
TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 295 3e-80
CB892890 weakly similar to GP|9758141|dbj| disease resistance pr... 293 2e-79
BG646612 weakly similar to GP|9758140|dbj| disease resistance pr... 289 3e-78
CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance prot... 268 6e-72
BF643326 weakly similar to GP|11994216|db disease resistance com... 204 3e-56
BG647778 weakly similar to PIR|T48468|T484 disease resistance-li... 186 4e-47
AW586679 similar to GP|9758140|dbj| disease resistance protein-l... 179 5e-45
TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum... 179 5e-45
TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust res... 139 5e-33
CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ163... 129 6e-30
BQ750499 similar to OMNI|NT01MC4764 transport protein HasD puta... 127 1e-29
BE319778 119 4e-27
TC89784 weakly similar to PIR|T05981|T05981 hypothetical protein... 100 3e-21
TC79479 similar to GP|12056928|gb|AAG48132.1 putative resistance... 88 1e-17
>TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (5%)
Length = 2535
Score = 1108 bits (2866), Expect(2) = 0.0
Identities = 556/602 (92%), Positives = 572/602 (94%)
Frame = +3
Query: 180 KTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLV 239
K P + VERIHQHCGY VPEFQSDEDAVN+LGLLLKKVEGSPLLLVLDDVWP SE LV
Sbjct: 648 KHPC*RLFVERIHQHCGYPVPEFQSDEDAVNRLGLLLKKVEGSPLLLVLDDVWPISEPLV 827
Query: 240 EKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLV 299
EK+QFQ++ FKILVTSRVAFPRFSTTCILKPLAHE+AVTLFHHYAQMEKNSSDII+KNLV
Sbjct: 828 EKIQFQISDFKILVTSRVAFPRFSTTCILKPLAHEEAVTLFHHYAQMEKNSSDIINKNLV 1007
Query: 300 EKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVL 359
EKVVRSC GLPLTIKVIATSLKNRP D+W KI KELSQGHSILDSNT+LLTRLQKIFDVL
Sbjct: 1008 EKVVRSCQGLPLTIKVIATSLKNRPRDMWRKIGKELSQGHSILDSNTDLLTRLQKIFDVL 1187
Query: 360 EDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVI 419
EDNP IMECFMD+ALFPEDHRIPVAALVDMWAELY+LDDNGIQAMEIINKLGIMNLANVI
Sbjct: 1188 EDNPTIMECFMDIALFPEDHRIPVAALVDMWAELYRLDDNGIQAMEIINKLGIMNLANVI 1367
Query: 420 IPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQ 479
IPRKNASDTDNNNYNNHFIILHDILRELGIYQS KEPFEQRKRLIID+N NKSGLAEKQQ
Sbjct: 1368 IPRKNASDTDNNNYNNHFIILHDILRELGIYQSAKEPFEQRKRLIIDINKNKSGLAEKQQ 1547
Query: 480 GLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYS 539
GLMT I SKFMRLCVK+NPQQL ARILSVS DETCA DWS M+PAQVEVLILN+HTKQYS
Sbjct: 1548 GLMTCILSKFMRLCVKRNPQQLTARILSVSADETCAFDWSQMEPAQVEVLILNLHTKQYS 1727
Query: 540 LPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLKKL 599
LPEWIAKMSKLRVLIITNY FHPSKLNNIELLGSLQNLERIRLERI VPSFGTLKNLKKL
Sbjct: 1728 LPEWIAKMSKLRVLIITNYDFHPSKLNNIELLGSLQNLERIRLERIYVPSFGTLKNLKKL 1907
Query: 600 SLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKL 659
SLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVL GICDI LKKL VTNCHKL
Sbjct: 1908 SLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLPIGICDIFLLKKLRVTNCHKL 2087
Query: 660 SSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN 719
SSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN
Sbjct: 2088 SSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN 2267
Query: 720 LKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNW 779
LKNLDMA+CASIELPFSVVNLQNLKTITCDEETAATWEDFQHML NMKIEVLHVDVNLNW
Sbjct: 2268 LKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNW 2447
Query: 780 LL 781
LL
Sbjct: 2448 LL 2453
Score = 251 bits (641), Expect(2) = 0.0
Identities = 141/185 (76%), Positives = 147/185 (79%), Gaps = 25/185 (13%)
Frame = +1
Query: 4 ATTHIITLTTRF--NDVLNKISEIVETARKNQSSRLLFRLILKDLSPVVQDIKHYNEHLD 61
A THIITLTTRF NDVLNKIS+I++ A+KNQSSR RLILKDLSPVVQDIK YN+HLD
Sbjct: 43 AATHIITLTTRFQFNDVLNKISQILQKAQKNQSSRRSLRLILKDLSPVVQDIKQYNDHLD 222
Query: 62 QPREEINSLIEE-----SACKSSFENDSY--VVDENQSLIVNDVEETLYKAREILELLNH 114
PREEINSLIEE SAC S ENDSY VV+ENQSL VNDVEETLYK REILELLNH
Sbjct: 223 HPREEINSLIEENDAGESACTCSSENDSYGYVVEENQSLTVNDVEETLYKTREILELLNH 402
Query: 115 E----------------NPKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATK 158
E N KFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATK
Sbjct: 403 EFNEDGQLFKRPFNVPENQKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATK 582
Query: 159 LCWDE 163
LCWDE
Sbjct: 583 LCWDE 597
Score = 52.0 bits (123), Expect = 9e-07
Identities = 24/28 (85%), Positives = 26/28 (92%)
Frame = +2
Query: 163 EEVNGKFKENIIFVTFSKTPMLKTIVER 190
+EVNGKFKENIIFVTFSKTPMLKTI +
Sbjct: 596 KEVNGKFKENIIFVTFSKTPMLKTICRK 679
Score = 31.2 bits (69), Expect = 1.6
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = -3
Query: 672 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFG 715
L L++S L +P S GKL L ++S C +S P E G
Sbjct: 2299 LAQLAMSRFFRLHKLPNSSGKLERLMQFEMSRCFRFNSFPMEVG 2168
>TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (7%)
Length = 2417
Score = 467 bits (1201), Expect(2) = 0.0
Identities = 237/336 (70%), Positives = 273/336 (80%), Gaps = 2/336 (0%)
Frame = +3
Query: 308 GLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIME 367
GLPL IKVIATS + RP++LW KIVKELS+G SILDSNTELL RLQKI DVLEDN E
Sbjct: 6 GLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISKE 185
Query: 368 CFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASD 427
CFMDLALFPED RIPVAAL+DMWAELY LDD+G +AM+IINKL MNLANV+I RKNASD
Sbjct: 186 CFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASD 365
Query: 428 TDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNK--SGLAEKQQGLMTRI 485
T+N YNNHFI+LHD+LRELG YQ+T+EP EQRKR +I+ N +K L EKQQG M I
Sbjct: 366 TENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHI 545
Query: 486 FSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIA 545
SK + K PQ++ AR LS+S DETCA DWS +QPA EVLILN+ TKQY+ PE +
Sbjct: 546 LSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELME 725
Query: 546 KMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCN 605
KM+KL+ LI+ N+G PS+LNN+ELL SL NL+RIRLERISVPSFGTLKNLKKLSLYMCN
Sbjct: 726 KMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMCN 905
Query: 606 TILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTG 641
T LAFEKGSILISD FPNLE+L++DYCKD+ L G
Sbjct: 906 TRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNG 1013
Score = 226 bits (577), Expect(2) = 0.0
Identities = 110/140 (78%), Positives = 125/140 (88%)
Frame = +1
Query: 641 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLD 700
G+CDIISLKKL++TNCHKLS LPQ+IGKLENLELLSL SCTDL +P SIG+L NL+ LD
Sbjct: 1012 GVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLD 1191
Query: 701 ISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQ 760
ISNCISLSSLPE+FGNLCNL+NLDM +CAS ELPFSVVNLQNLK +TCDEETAA+WE FQ
Sbjct: 1192 ISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLK-VTCDEETAASWESFQ 1368
Query: 761 HMLHNMKIEVLHVDVNLNWL 780
M+ N+ IEV HV+VNLNWL
Sbjct: 1369 SMISNLTIEVPHVEVNLNWL 1428
Score = 29.6 bits (65), Expect = 4.6
Identities = 17/45 (37%), Positives = 24/45 (52%)
Frame = -1
Query: 672 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 716
L L +S L +P S GKL L L++S+ + SLP E G+
Sbjct: 1280 LAQLVMSRFRRLHRLPKSSGKLERLMQLEMSSSLRFESLPIESGS 1146
>BG646517
Length = 668
Score = 405 bits (1041), Expect = e-113
Identities = 200/220 (90%), Positives = 210/220 (94%)
Frame = +2
Query: 159 LCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKK 218
LCWD+EVNGKFKENIIFVTFSKTPMLKTIVERI +HCGY VPEFQSDEDAVNKL LLLKK
Sbjct: 2 LCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKK 181
Query: 219 VEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVT 278
VEGSPLLLVLDDVWP+SESLV+KLQF+++ FKILVTSRV+FPRF TTCILKPLAHEDAVT
Sbjct: 182 VEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVT 361
Query: 279 LFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQG 338
LFHHYAQMEKNSSDIIDKNLVEKVVRSC GLPLTIKVIATSL+NRP DLW KIV ELSQG
Sbjct: 362 LFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQG 541
Query: 339 HSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPED 378
HSILDSNTELLTRLQKIFDVLEDNP IMECFMD+ALFPED
Sbjct: 542 HSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPED 661
>TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (45%)
Length = 2140
Score = 327 bits (839), Expect = 8e-90
Identities = 191/565 (33%), Positives = 324/565 (56%), Gaps = 8/565 (1%)
Frame = +3
Query: 224 LLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHY 283
+L++LDDVW S S++E+L F+M K +V SR FP F+ T ++ L +DA++LF H+
Sbjct: 396 ILVILDDVW--SPSVLEQLVFRMPNCKFIVVSRFIFPIFNATYKVELLDKDDALSLFCHH 569
Query: 284 AQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILD 343
A +K+ ++NLV++VV C LPL +KVI SL+++ W + LSQG SI +
Sbjct: 570 AFGQKSIPFAANQNLVKQVVAECGNLPLALKVIGASLRDQNEMFWLSVKTRLSQGLSIDE 749
Query: 344 S-NTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQ 402
S L+ R+ + L + KI ECF+DL FPED +IP+ L++MW E++ + + +
Sbjct: 750 SYERNLIDRMAISTNYLPE--KIKECFLDLCSFPEDKKIPLEVLINMWVEIHDIHET--E 917
Query: 403 AMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHF---IILHDILRELGIYQSTKEPFEQ 459
A I+ +L NL ++ + Y++ F + HDILR+L + S + Q
Sbjct: 918 AYAIVVELSNKNLLTLVEEARAGG-----MYSSCFEISVTQHDILRDLALNLSNRGNINQ 1082
Query: 460 RKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWS 519
R+RL++ + L ++ ++ Q A+I+S+ T E DW
Sbjct: 1083RRRLVMPKREDNGQLPKEW---------------LRYADQPFEAQIVSIHTGEMRKSDWC 1217
Query: 520 HMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLER 579
+++ + EVLI+N + +Y LP +I +M KLR L++ N+ + L+NI + +L NL
Sbjct: 1218NLEFPKAEVLIINFTSSEYFLPPFINRMPKLRALMVINHSTSYACLHNISVFKNLTNLRS 1397
Query: 580 IRLERISVPSFG--TLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVV 637
+ E++S+P +++L+KL + +C + E I+D FPN+ EL +D+C+D+
Sbjct: 1398LWFEKVSIPHLSGIVMESLRKLFIVLCKINNSLEGKDSNIADIFPNISELTLDHCEDVTE 1577
Query: 638 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 697
L + IC I SL+ L++TNCH L+ LP ++G L LE+L L +C +L +P SI + LK
Sbjct: 1578LPSSICRIQSLQNLSLTNCHSLTRLPIELGSLRYLEILRLYACPNLRTLPPSICGMTRLK 1757
Query: 698 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLKTI-TCDEETAAT 755
++DI+ C+ L+S P+ G L NL+ +DM C I +P S ++L ++ CD+E +
Sbjct: 1758YIDIAQCVYLASFPDAIGKLVNLEKIDMRECPMITNIPKSALSLNFFCSL*ICDDEVSWM 1937
Query: 756 WEDFQHMLHNMKIEVLHVDVNLNWL 780
W++ Q + N+ I+V+ ++ +L+WL
Sbjct: 1938WKEVQKVKLNVDIQVVEIEYDLDWL 2012
Score = 53.9 bits (128), Expect = 2e-07
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Frame = +2
Query: 116 NPKFTVGLDIPFSKLKMELLRGGSSTLV--LTGLGGLGKTTLATKLCWDEEVNGKFKENI 173
N + GL+ +K+ ME++ G + ++G+GG GKTTLA ++C DE+V G FKE I
Sbjct: 71 NLDLSFGLEFGKNKV-MEMVVGRKDFCLVGISGIGGSGKTTLAREICRDEQVRGYFKERI 247
Query: 174 IFVTFSKTP 182
+F+T S++P
Sbjct: 248 LFLTVSQSP 274
>TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (12%)
Length = 1137
Score = 308 bits (789), Expect = 5e-84
Identities = 164/307 (53%), Positives = 215/307 (69%), Gaps = 4/307 (1%)
Frame = +1
Query: 478 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 537
QQG+++ +S + VKQ ++AARIL +STDE + DW MQP + EVL+LN+ + Q
Sbjct: 25 QQGIISNWYSFITGMLVKQKQLKVAARILCISTDEIFSSDWCDMQPDKAEVLVLNLRSDQ 204
Query: 538 YSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLK 597
YSLP++ KM KL+VLI+TNYGF S+L ELLGSL NL+RIRLE++SVP LKNL+
Sbjct: 205 YSLPDFTKKMRKLKVLIVTNYGFSRSELTKFELLGSLSNLKRIRLEKVSVPCLCILKNLR 384
Query: 598 KLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCH 657
KLSL+MC+T AFE SI ISDA PNL EL+IDYC DL+ L C I +LKKL++TNCH
Sbjct: 385 KLSLHMCSTNNAFESCSIQISDAMPNLVELSIDYCNDLIKLPGEFCKITTLKKLSITNCH 564
Query: 658 KLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNL 717
K S++PQDIGKL NLE+L L SC+DL+ IP S+ L L+ LDIS+C++L LP GNL
Sbjct: 565 KFSAMPQDIGKLVNLEVLRLCSCSDLKEIPESVADLNKLRCLDISDCVTLHILPNNIGNL 744
Query: 718 CNLKNLDMATCASI-ELPFSVVNLQNLK---TITCDEETAATWEDFQHMLHNMKIEVLHV 773
L+ L M C+++ ELP SV+N NLK + CDEE +A WE + + +KI + V
Sbjct: 745 QKLEKLYMKGCSNLSELPDSVINFGNLKHEMQVICDEEGSALWEHLSN-IPKLKIYMPKV 921
Query: 774 DVNLNWL 780
+ NL WL
Sbjct: 922 EHNLIWL 942
>TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (5%)
Length = 1294
Score = 303 bits (775), Expect = 2e-82
Identities = 156/275 (56%), Positives = 204/275 (73%), Gaps = 4/275 (1%)
Frame = +3
Query: 510 TDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIE 569
TDE+ + DW MQP +VEVL+LN+ + QYSLP++ KMSKL+VLI+TNYGFH S+L E
Sbjct: 120 TDESFSSDWCDMQPDEVEVLVLNLQSDQYSLPDFTDKMSKLKVLIVTNYGFHRSELIKFE 299
Query: 570 LLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNI 629
LLG L NL+RIRLE++SVP LKNL+KLSL+MCNT AFE SI ISDA PNL EL+I
Sbjct: 300 LLGFLSNLKRIRLEKVSVPCLSILKNLQKLSLHMCNTRDAFENYSIQISDAMPNLVELSI 479
Query: 630 DYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTS 689
DYC DL+ L G +I +LKK+++TNCHKLS++PQDI KLENLE+L L SC+DL I S
Sbjct: 480 DYCNDLIKLPDGFSNITTLKKISITNCHKLSAIPQDIEKLENLEVLRLCSCSDLVEISES 659
Query: 690 IGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK---T 745
+ L L+ DIS+C+SLS LP + G+L L+ M C+++ ELP+SV+NL N+K
Sbjct: 660 VSGLNKLRCFDISDCVSLSKLPNDIGDLKKLEKFYMKGCSNLSELPYSVINLGNVKHEIH 839
Query: 746 ITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
+ CDEE AA WE F + + N+KI++ V++NLNWL
Sbjct: 840 VICDEEGAALWEHFPN-IPNLKIDMPKVEINLNWL 941
>TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (16%)
Length = 1128
Score = 295 bits (756), Expect = 3e-80
Identities = 165/351 (47%), Positives = 226/351 (64%), Gaps = 6/351 (1%)
Frame = +1
Query: 436 HFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVK 495
H++ H +LR+L I +++E ++R RLIID + N
Sbjct: 1 HYVTQHGLLRDLAIRDNSQEAEDKRNRLIIDTSANN-----------------LPSWWTS 129
Query: 496 QNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLII 555
+N + ARILS+STDE W ++QP +VEVL+LN+ K+++LP ++ KM KL+ LII
Sbjct: 130 ENEYHIKARILSISTDEIFTSKWCNLQPTEVEVLVLNLREKKFALPMFMKKMKKLKALII 309
Query: 556 TNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFG----TLKNLKKLSLYMCNTILAFE 611
TNY F P++L N E+L L NL+RIRLE++SVP G LKNL+K S +MCN AFE
Sbjct: 310 TNYDFFPAELENFEVLDHLSNLKRIRLEKVSVPFLGKTVLQLKNLQKCSFFMCNVNKAFE 489
Query: 612 KGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLEN 671
+I S+ PNL E+N DYC D+V L I +I+SLKKL++TNCHKL +L + IGKL N
Sbjct: 490 NCTIEDSEMLPNLMEMNFDYC-DMVELPNVISNIVSLKKLSITNCHKLCALHEGIGKLVN 666
Query: 672 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI 731
LE L LSSC+ L +P SI L LK L+IS CISLS LPE G L L+ L+M C++I
Sbjct: 667 LESLRLSSCSGLLELPNSIRNLHVLKFLNISGCISLSQLPENIGELEKLEKLNMRGCSNI 846
Query: 732 -ELPFSVVNLQNLKTITC-DEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
ELP SV+ L+ LK + C DEETA WE ++++L ++K+EV+ D NLN+L
Sbjct: 847 SELPSSVMELEGLKHVVCGDEETAEKWEPYKNILGDLKVEVVQEDFNLNFL 999
>CB892890 weakly similar to GP|9758141|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 852
Score = 293 bits (750), Expect = 2e-79
Identities = 155/305 (50%), Positives = 208/305 (67%), Gaps = 3/305 (0%)
Frame = +2
Query: 353 QKIFD-VLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLG 411
QK+ + L+DN + ECFMDL LF ED +IPVAAL+DMW ELY LDD+ I+ M I+ KL
Sbjct: 2 QKVLENALQDNHIVKECFMDLGLFFEDKKIPVAALIDMWTELYDLDDDNIKGMNIVRKLA 181
Query: 412 IMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNK 471
+L +++ R+ + D+ YN+HF+ HD+L+E+ I+Q+ +EPFE RKRLI D+N N
Sbjct: 182 NWHLVKLVVSREVTTHVDHY-YNHHFLTQHDLLKEISIHQARQEPFELRKRLIFDVNENS 358
Query: 472 SGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPA-QVEVLI 530
+QN Q AR LS+S D+ DWS+++ QVEVLI
Sbjct: 359 WD---------------------QQNQQNTIARTLSISPDKILTSDWSNVEKIKQVEVLI 475
Query: 531 LNIHTKQYSLPEWIAKMSKLRVLIITNY-GFHPSKLNNIELLGSLQNLERIRLERISVPS 589
LN+HT++Y+LPE I KM+KL+VLIITNY GFH ++L+N E+LG L NL RIRL ++SVPS
Sbjct: 476 LNLHTEKYTLPECIKKMTKLKVLIITNYKGFHCAELDNFEILGCLPNLRRIRLHQVSVPS 655
Query: 590 FGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLK 649
L NL+KLSLY C T AF+ ++ ISD PNL+EL +DYCKDLV L +G+CDI SLK
Sbjct: 656 LCKLVNLRKLSLYFCETKQAFQSNTVSISDILPNLKELCVDYCKDLVTLPSGLCDITSLK 835
Query: 650 KLNVT 654
KL++T
Sbjct: 836 KLSIT 850
>BG646612 weakly similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (7%)
Length = 735
Score = 289 bits (739), Expect = 3e-78
Identities = 170/243 (69%), Positives = 183/243 (74%), Gaps = 30/243 (12%)
Frame = +2
Query: 4 ATTHI-ITLTTRFNDVLNKISEIVETARKNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQ 62
A HI ITLTTRF+DVLNKIS+IVETARK+QS+R L R ILKDL+ VVQDIK YNEHLD
Sbjct: 5 APLHIFITLTTRFHDVLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHLDH 184
Query: 63 PREEINSLIEE-----SACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNHE-- 115
PREEI +L+EE SACK S ENDSYVVDENQSLIVNDVEETLYKAREILELLN+E
Sbjct: 185 PREEIKTLLEENDAEESACKCSSENDSYVVDENQSLIVNDVEETLYKAREILELLNYETF 364
Query: 116 -------------------NPKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLA 156
NPKFTVGLDIPFSKLKMEL+R GSSTLVLTGLGGLGKTTLA
Sbjct: 365 EHKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLA 544
Query: 157 TKLCWDEEVNGKFKEN-IIFVTFSKTPMLKTIV-ERIHQHCGYSVP-EFQSDEDAVNKLG 213
TKLCWD+EVNGKFKEN I K P LKT E+ CGY VP F+ DE+AVNKL
Sbjct: 545 TKLCWDQEVNGKFKENHYICHPSQKHPWLKTYCREK*LNICGYPVP*VFKVDENAVNKLE 724
Query: 214 LLL 216
LLL
Sbjct: 725 LLL 733
>CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance protein - rice
(fragment), partial (8%)
Length = 706
Score = 268 bits (685), Expect = 6e-72
Identities = 137/190 (72%), Positives = 155/190 (81%)
Frame = -1
Query: 235 SESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDII 294
SE LVEK +FQ++ +KILVT VAF RF T ILKPLA ED+VTLF HY ++EKNSS I
Sbjct: 703 SEDLVEKFKFQISDYKILVTQGVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIP 524
Query: 295 DKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQK 354
DK+L+EKVV C GLPL IKVIATS + RP++LW KIVKELS+G SILDSNTELL RLQK
Sbjct: 523 DKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQK 344
Query: 355 IFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMN 414
I DVLEDN ECFMDLALFPED RIPVAAL+DMWAELY LDD+G +AM+IINKL MN
Sbjct: 343 ILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMN 164
Query: 415 LANVIIPRKN 424
LANV+I R N
Sbjct: 163 LANVLIARYN 134
>BF643326 weakly similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (2%)
Length = 486
Score = 204 bits (519), Expect(2) = 3e-56
Identities = 102/125 (81%), Positives = 112/125 (89%)
Frame = +2
Query: 585 ISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICD 644
ISVPSFGT+KNLKKLSLYMCNT LAFEKGSILISD FPNLE+L+IDY KD+V L G+CD
Sbjct: 2 ISVPSFGTMKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSIDYSKDMVALPNGVCD 181
Query: 645 IISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNC 704
I SLKKL++TNCHKLSSLPQDIGKL NLELLSL SCTDL +P SIG+LLNL+ LDISNC
Sbjct: 182 IASLKKLSITNCHKLSSLPQDIGKLMNLELLSLISCTDLVELPDSIGRLLNLRLLDISNC 361
Query: 705 ISLSS 709
ISLSS
Sbjct: 362 ISLSS 376
Score = 33.5 bits (75), Expect(2) = 3e-56
Identities = 13/18 (72%), Positives = 17/18 (94%)
Frame = +3
Query: 710 LPEEFGNLCNLKNLDMAT 727
LPE+FGNLCNL+NL M++
Sbjct: 378 LPEDFGNLCNLRNLYMSS 431
>BG647778 weakly similar to PIR|T48468|T484 disease resistance-like protein -
Arabidopsis thaliana, partial (4%)
Length = 660
Score = 186 bits (471), Expect = 4e-47
Identities = 105/201 (52%), Positives = 131/201 (64%), Gaps = 3/201 (1%)
Frame = +2
Query: 117 PKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFV 176
P TVGLD P +LK LL G S LVLTGL G GKTTLAT LCWD++V GKF ENI+F
Sbjct: 20 PDLTVGLDFPLIQLKSWLLGSGVSVLVLTGLAGSGKTTLATLLCWDDKVRGKFGENILFF 199
Query: 177 TFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPLLLVLDDVWPSS 235
T SK P LK IV+ + +HCG+ P D+DAV L LL K+ E P++LVLD+V P S
Sbjct: 200 TVSKIPNLKNIVQTLFRHCGHDEPCLIDDDDAVKHLRSLLTKIGESCPMMLVLDNVCPGS 379
Query: 236 ESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA--QMEKNSSDI 293
ESLVE Q Q+ KIL+TSRV FPRFS + LKPL +DAVTLF +A ++ +
Sbjct: 380 ESLVEDFQVQVPDCKILITSRVEFPRFS-SLFLKPLRDDDAVTLFGSFALPNDATRATYV 556
Query: 294 IDKNLVEKVVRSCNGLPLTIK 314
+ V++V + C G PL +K
Sbjct: 557 PAEKYVKQVAKGCWGSPLALK 619
>AW586679 similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 481
Score = 179 bits (453), Expect = 5e-45
Identities = 88/159 (55%), Positives = 115/159 (71%)
Frame = +3
Query: 147 LGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDE 206
LGG GKTTLA KLCW+++ GKF NI FVT S+TP LK IV+ + + CG VP+F ++E
Sbjct: 3 LGGSGKTTLAKKLCWEQDFKGKFGGNIFFVTVSETPNLKNIVKTLFEQCGRPVPDFTNEE 182
Query: 207 DAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTC 266
DA+N+LG LL + E S +LLVLDDVWP SESLVEK F++ +KILVTSRV F RF T C
Sbjct: 183 DAINQLGHLLSQFERSQILLVLDDVWPGSESLVEKFTFKLPDYKILVTSRVGFRRFGTPC 362
Query: 267 ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRS 305
L PL + A +LF HYAQ+ S + D +LV+++V++
Sbjct: 363 QLNPLDEDPAASLFRHYAQLHHIISYMPDGDLVDEIVKA 479
>TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum glaucum},
partial (10%)
Length = 827
Score = 179 bits (453), Expect = 5e-45
Identities = 95/167 (56%), Positives = 122/167 (72%), Gaps = 9/167 (5%)
Frame = +2
Query: 77 KSSFENDSYVVDENQSLIVNDVEETLYKAREI----LELLNH-----ENPKFTVGLDIPF 127
K +F +DS+ V ++ + +E L+ ++I + + ENP+FTVGLD
Sbjct: 308 KKNFGSDSHAVAHDKPKTFYEFKEILFFDKKISGYGVSIFKGPFGVPENPEFTVGLDSQL 487
Query: 128 SKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTI 187
K KMELLR G STL+LTGLGG+GKTTLATKLC D++V GKFKENIIFVTFSKTPMLK +
Sbjct: 488 IKPKMELLRDGRSTLLLTGLGGMGKTTLATKLCLDQQVKGKFKENIIFVTFSKTPMLKIM 667
Query: 188 VERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPS 234
V+R+ +H GY VPE+QSDE AVN +G LL+K+EGSP+L VLDDV P+
Sbjct: 668 VQRLFEHGGYPVPEYQSDEXAVNGMGNLLRKIEGSPILXVLDDVGPA 808
>TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust resistance
protein {Oryza sativa (japonica cultivar-group)},
partial (1%)
Length = 954
Score = 139 bits (349), Expect = 5e-33
Identities = 81/165 (49%), Positives = 104/165 (62%), Gaps = 19/165 (11%)
Frame = +2
Query: 88 DENQSLIVNDVEETLYKAREILELLNHEN-------------------PKFTVGLDIPFS 128
D+++ + DVE K RE+L++L+ EN P+FTVGLD+
Sbjct: 464 DDDKQKMGKDVEG---KLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLI 634
Query: 129 KLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIV 188
KLK+E+LR G STL+LTGLGG+GKTTLATKLC D++V GKFKENIIFVTFSK P + +
Sbjct: 635 KLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKHPC*RLL* 814
Query: 189 ERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWP 233
+ + N LGLLL+K+EGSP+LLVLDDVWP
Sbjct: 815 RDYLNIADIRCLSTKVMKMRENGLGLLLRKIEGSPILLVLDDVWP 949
>CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ1637 -
Methanococcus jannaschii, partial (2%)
Length = 473
Score = 129 bits (323), Expect = 6e-30
Identities = 68/90 (75%), Positives = 71/90 (78%), Gaps = 16/90 (17%)
Frame = +2
Query: 95 VNDVEETLYKAREILELLNHE----------------NPKFTVGLDIPFSKLKMELLRGG 138
VN+VEETLYK +EILELLNHE N KFTVGLDIPF+KLKMELLRGG
Sbjct: 5 VNEVEETLYKTKEILELLNHEFNEDGQLFKRPFNVPENQKFTVGLDIPFTKLKMELLRGG 184
Query: 139 SSTLVLTGLGGLGKTTLATKLCWDEEVNGK 168
SSTLVLTGL GLGKTTLATKLCWDEEVNGK
Sbjct: 185 SSTLVLTGLRGLGKTTLATKLCWDEEVNGK 274
>BQ750499 similar to OMNI|NT01MC4764 transport protein HasD putative
{Magnetococcus sp. MC-1}, partial (3%)
Length = 800
Score = 127 bits (320), Expect = 1e-29
Identities = 85/219 (38%), Positives = 118/219 (53%), Gaps = 49/219 (22%)
Frame = +1
Query: 43 LKDLSPVVQDIKHYNEHLDQPREEINSLI------EESACKSS------------FENDS 84
L +L+P+V++IK YN+ LD+P EEI L EE KS ++
Sbjct: 145 LNNLAPLVKEIKVYNDFLDRPSEEIERLEKHIREGEELVRKSKKLTLWNFLSFPGYQGKL 324
Query: 85 YVVDEN--QSLIVN-------DVEETLYKAREILELLN---------------------- 113
DE + L VN D+ + + K EI ++
Sbjct: 325 KKKDEGLQRHLSVNVQLENKKDLIKLVAKVDEISKIFEILMRKENLGQFDGSQIRGLCGA 504
Query: 114 HENPKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENI 173
E P+ +G+D P +KLK++LL+ G S LVLTGLGG GK+TLA KLCW+ ++ GKF NI
Sbjct: 505 PEEPQ-CMGVDEPLNKLKIQLLKDGVSVLVLTGLGGSGKSTLAKKLCWNPQIKGKFGGNI 681
Query: 174 IFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKL 212
FVT SKTP LK IV+ + +HCG VPEFQ+DEDA+ ++
Sbjct: 682 FFVTVSKTPNLKNIVQTLFEHCGLRVPEFQTDEDAITRV 798
>BE319778
Length = 378
Score = 119 bits (298), Expect = 4e-27
Identities = 57/60 (95%), Positives = 58/60 (96%)
Frame = +2
Query: 422 RKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGL 481
RKNASDTDNNNYNNHFIILHDILRELGIYQS KEPFEQRKRLIID+N NKSGLAEKQQGL
Sbjct: 197 RKNASDTDNNNYNNHFIILHDILRELGIYQSAKEPFEQRKRLIIDINKNKSGLAEKQQGL 376
Score = 41.6 bits (96), Expect = 0.001
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +1
Query: 403 AMEIINKLGIMNLANVIIPR 422
AMEIINKLGIMNLANVIIPR
Sbjct: 1 AMEIINKLGIMNLANVIIPR 60
>TC89784 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (20%)
Length = 1100
Score = 100 bits (248), Expect = 3e-21
Identities = 70/205 (34%), Positives = 115/205 (55%), Gaps = 11/205 (5%)
Frame = +1
Query: 113 NHENPKFTVGLDIPFSKLKMELLRGGSSTLV--LTGLGGLGKTTLATKLCWDEEVNGKFK 170
N+ N +VGLD+ K+K E++ G V + G+GG GKTTL ++C DE+V F
Sbjct: 517 NYGNLSLSVGLDLGKKKVK-EMVMGREDLWVVGIHGIGGSGKTTLVKEICKDEQVRCYFN 693
Query: 171 ENIIFVTFSKTPMLKTIVERIHQHC--------GYSVPEFQSDEDAVNKLGLLLKKVEGS 222
E I+F+T S++P ++ + +I H Y VP + + ++ +
Sbjct: 694 EKILFLTVSQSPNVEQLRSKIWGHIMGNRNLNPNYVVPRWIPQFECRSE----------A 843
Query: 223 PLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPR-FSTTCILKPLAHEDAVTLFH 281
L+VLDDVW S++++E+L ++ G K +V SR FP FS T ++ L+ EDA++LF
Sbjct: 844 RTLVVLDDVW--SQAVLEQLVCRIPGCKFVVVSRFQFPTIFSATYKVELLSEEDALSLFC 1017
Query: 282 HYAQMEKNSSDIIDKNLVEKVVRSC 306
H+A +K+ ++NLV++VV C
Sbjct: 1018HHAFGQKSIPLTANENLVKQVVSEC 1092
>TC79479 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (21%)
Length = 1087
Score = 87.8 bits (216), Expect = 1e-17
Identities = 66/201 (32%), Positives = 103/201 (50%)
Frame = +1
Query: 546 KMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCN 605
KM+ L+ LII + F N L SL+ LE S+PS+ K L L L +
Sbjct: 97 KMTGLQTLIIRSLCFAEGPKN---LPNSLRVLEWWGYPSQSLPSYFYPKKLAVLKLPHSS 267
Query: 606 TILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQD 665
F + S F N+ LN D CK ++ + +L++L++ +C L +
Sbjct: 268 ----FMSLELSKSKKFVNMTLLNFDECK-IITHIPDVSGAPNLERLSLDSCENLVEIHDS 432
Query: 666 IGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDM 725
+G L+ LE+L+L SC L +P L +L+HL++S+C SL S PE GN+ N+ +L +
Sbjct: 433 VGFLDKLEILNLGSCAKLRNLPPI--HLTSLQHLNLSHCSSLVSFPEILGNMKNITSLSL 606
Query: 726 ATCASIELPFSVVNLQNLKTI 746
A E P+S+ NL LK++
Sbjct: 607 EYTAIREFPYSIGNLPRLKSL 669
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,654,434
Number of Sequences: 36976
Number of extensions: 352864
Number of successful extensions: 2494
Number of sequences better than 10.0: 330
Number of HSP's better than 10.0 without gapping: 2146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2324
length of query: 781
length of database: 9,014,727
effective HSP length: 104
effective length of query: 677
effective length of database: 5,169,223
effective search space: 3499563971
effective search space used: 3499563971
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135229.3


