

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135160.10 + phase: 0
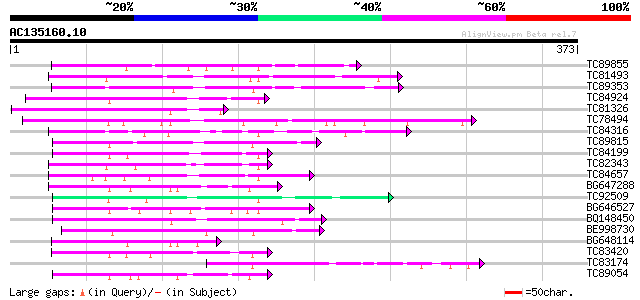
(373 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89855 83 2e-16
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 76 2e-14
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 74 1e-13
TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein... 70 2e-12
TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein... 69 3e-12
TC78494 69 3e-12
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 68 5e-12
TC89815 67 1e-11
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 67 1e-11
TC82343 similar to GP|10177628|dbj|BAB10775. gb|AAF30317.1~gene_... 65 4e-11
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 65 5e-11
BG647288 weakly similar to GP|5903055|gb|A May be a pseudogene o... 63 2e-10
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 62 3e-10
BG646527 61 6e-10
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 58 5e-09
BE998730 57 1e-08
BG648114 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.... 55 3e-08
TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pse... 55 6e-08
TC83174 55 6e-08
TC89054 weakly similar to GP|5903055|gb|AAD55614.1| May be a pse... 54 7e-08
>TC89855
Length = 766
Score = 82.8 bits (203), Expect = 2e-16
Identities = 69/229 (30%), Positives = 106/229 (46%), Gaps = 25/229 (10%)
Frame = +3
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATR-RNRLLILQNA----------- 75
+++ IL +LP K L RF CV+KSW+++ ++P F RN L+
Sbjct: 69 DDIVFSILSKLPIKHLKRFACVRKSWSHLFENPIFMNMFRNNLVSKSQTGYDDDDACLIC 248
Query: 76 -----PNMKFIFCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFC--LKGISSCITRHD 128
P K F G +K I + L PQ + + + C + GI D
Sbjct: 249 HWVLDPVKKLSFLTGEKFEKEIKL-DLPPQVQIQQNDFLDYISILCSAINGILCIYNWFD 425
Query: 129 --QLILWNPTTKEVHLIPR--APSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMA 182
Q++LWNPTT EVH++P SL N + D+ LYGFG +DD+KV+++ + M
Sbjct: 426 PSQIVLWNPTTNEVHVVPSNLPESLPNVFVDQFLYGFGYDHDSDDYKVIRV-VRFREDMF 602
Query: 183 KINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWI 231
K + +IY L + SW L PI + Y ++GV +W+
Sbjct: 603 KTHDPF-YEIYSLRSHSWRKLDVDIPIVFYGPLSSEVY---LDGVCHWL 737
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 76.3 bits (186), Expect = 2e-14
Identities = 65/251 (25%), Positives = 111/251 (43%), Gaps = 18/251 (7%)
Frame = +1
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF-----ATRRNRLLILQNAPNMKF 80
L E+ EIL R+P K L R + K W N+I S F + R+ ++IL+ +
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKSRDSVIILRQHSRLYE 195
Query: 81 IFCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEV 140
+ + + K + + + RI++ GSC+G+ C+ I+ D + WNPT ++
Sbjct: 196 LDLNSMDRVKELDHPLMCYSN--RIKVLGSCNGLLCICNIA------DDIAFWNPTIRKH 351
Query: 141 HLIPRAPSLGNHYSDES---------LYGFG--AVNDDFKVVKLNISNSNRMAKINSLLK 189
+IP P + ++ + +YGFG + DD+K+V +ISN + +
Sbjct: 352 RIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLV--SISNFVDLHNRSYDSH 525
Query: 190 ADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAAR--ILCFDF 247
IY + + W PL P R V+G +W+ +R I+ FD
Sbjct: 526 VTIYTMGSDVWMPLPGVPYALCCAR----PMGVFVSGALHWVVPRALEPDSRDLIVAFDL 693
Query: 248 RDNQFRKLEAP 258
R FR++ P
Sbjct: 694 RFEVFREVALP 726
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 73.6 bits (179), Expect = 1e-13
Identities = 70/251 (27%), Positives = 105/251 (40%), Gaps = 19/251 (7%)
Frame = +3
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGN 87
EE+ +EILLRLP +SL +F+CV K W +I P FA + + I P + +F
Sbjct: 111 EEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFA--KKHVSISTAYPQLVSVFVSIAK 284
Query: 88 -DQKSIPIKSLFPQDVA-RIE-------------IYGSCDGVFCLKGISSCITRHDQLIL 132
+ S P+K L A R+E I GSC+G+ CL Q L
Sbjct: 285 CNLVSYPLKPLLDNPSAHRVEPADFEMIHTTSMTIIGSCNGLLCLSDFY-------QFTL 443
Query: 133 WNPTTK-EVHLIPRAPSLGNHYSDESLY---GFGAVNDDFKVVKLNISNSNRMAKINSLL 188
WNP+ K + P + + S LY G+ VND +KV+ + + N + + +L
Sbjct: 444 WNPSIKLKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAV-VQNCYNLDETKTL- 617
Query: 189 KADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFR 248
IY K WT + P + V+G WI S I+ FD
Sbjct: 618 ---IYTFGGKDWTTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIVSK-----KVIVFFDIE 773
Query: 249 DNQFRKLEAPK 259
+ ++ P+
Sbjct: 774 KETYGEMSLPQ 806
>TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein F8A12.9
[imported] - Arabidopsis thaliana, partial (9%)
Length = 575
Score = 69.7 bits (169), Expect = 2e-12
Identities = 59/175 (33%), Positives = 79/175 (44%), Gaps = 14/175 (8%)
Frame = +1
Query: 11 FSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT------ 64
+S+ R + + M L +EL I+ LLRLP KSL FKC+ K W +II P+FA
Sbjct: 16 YSKQRVKNNAAQSMDLPQELIIQFLLRLPVKSLLVFKCICKLWFSIISDPHFANSHFQLN 195
Query: 65 ----RRNRLLILQNAPNMKFIFCDGG-NDQKSIP-IKSLFPQDVARIEIYGSCDGVFCLK 118
R L I +P ++ I D ND + P P EI GSC G +
Sbjct: 196 HAKHTRRFLCISALSPEIRSIDFDAFLNDAPASPNFNCSLPDSYFPFEIKGSCRGFIFM- 372
Query: 119 GISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVV 171
RH + +WNP+T I S N + +LYGFG DD+ VV
Sbjct: 373 ------YRHPNIYIWNPSTGSKRQI--LMSAFNTKAYINLYGFGYDQSRDDYVVV 513
>TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein F13I12.200
- Arabidopsis thaliana, partial (12%)
Length = 838
Score = 68.9 bits (167), Expect = 3e-12
Identities = 50/160 (31%), Positives = 73/160 (45%), Gaps = 17/160 (10%)
Frame = +3
Query: 2 CYLIVHNKKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPY 61
C + V+ S K K + L EL +ILLRLP KSL+RFK V+KSW ++I +P+
Sbjct: 96 CKIRVYKMVESSEEKKKKTTTLPYLPHELIFQILLRLPVKSLTRFKSVRKSWFSLISAPH 275
Query: 62 FATRRNRLLILQNAPNMKFIFCDGGNDQKSIPIKSLFPQDVAR--------------IEI 107
FA +L ++A + ++ +SI K+ D +EI
Sbjct: 276 FANSHFQLTSAKHAASRIMFISTLSHETRSIDFKAFLNDDDPASLNITFSLTRSHFPVEI 455
Query: 108 YGSCDGVFCLKGISSCITRHDQLILWNPTT---KEVHLIP 144
GSC G L R + +WNP+T K +HL P
Sbjct: 456 RGSCRGFILL-------YRPPDIYIWNPSTGFKKHIHLSP 554
>TC78494
Length = 1450
Score = 68.9 bits (167), Expect = 3e-12
Identities = 76/347 (21%), Positives = 143/347 (40%), Gaps = 48/347 (13%)
Frame = +1
Query: 9 KKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA----- 63
K + ++ +S L EL IL RLP++ L + K V K+W N+I +F
Sbjct: 136 KPVQKPKRNNVSTSMENLPHELVSNILSRLPSRELLKNKLVCKTWYNLITDSHFTNNYYS 315
Query: 64 ------TRRNRLLILQ-----NAPNMKFIFCDGGNDQKSIPIKSLF--PQDVAR-----I 105
+ LL+++ + + ND K SL P+
Sbjct: 316 FHNQLQNQEEHLLVIRRPFISSLKTTISLHSSTFNDPKKNVCSSLLNPPEQYNSEHKYWS 495
Query: 106 EIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNH----YSDESLYGF 161
EI G C+G++ L+G + +L N + ++ +P + ++ +D + +GF
Sbjct: 496 EIMGPCNGIYLLQG--------NPNVLMNASLQQFKALPESHLTDSNGIYSLTDYASFGF 651
Query: 162 GAVNDDFKVVKLN---ISNSNRMAKINSLLKADIYDLSTKSWTPLVSHP---PITMV--T 213
+D+KV+ L + ++ K ++Y L++ SW L + PI + +
Sbjct: 652 DPKTNDYKVIVLKDLWLKETDERQK--GYWTGELYSLNSNSWKKLDAETLPLPIEICGSS 825
Query: 214 RIQPSRYNTLVNGVYYWIT------SSDGSDAARILCFDFRDNQFRKLEAPKL--GHYIP 265
SR T VN +W + S G + +L FD + FRK++ P++
Sbjct: 826 SSSSSRVYTYVNNCCHWWSFVNNHDESQGMNQDFVLSFDIVNEVFRKIKVPRICESSQET 1005
Query: 266 FFCDDVFEIKGYLGYVVQ-YRCRIVWLEIWTL----EQNGWAKKYNI 307
F FE +G++V R + ++W + ++ W K+Y++
Sbjct: 1006FVTLAPFEESSTIGFIVNPIRGNVKHFDVWVMRDYWDEGSWIKQYSV 1146
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 68.2 bits (165), Expect = 5e-12
Identities = 71/260 (27%), Positives = 109/260 (41%), Gaps = 21/260 (8%)
Frame = +3
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDG 85
L EEL + ILLRLP +SL RFKCV KSW + +FA N LI P + + C+
Sbjct: 42 LPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFA--NNHFLISTVYPQL--VACES 209
Query: 86 GN-----DQKSIPIKSLFPQDVA-----------RIEIYGSCDGVFCL-KGISSCITRHD 128
+ + K+ PI+SL R I GSC+G CL C+
Sbjct: 210 VSAYRTWEIKTYPIESLLENSSTTVIPVSNTGHQRYTILGSCNGFLCLYDNYQRCVR--- 380
Query: 129 QLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMAKINS 186
LWNP+ ++L ++ + + YGFG VN +K++ + +
Sbjct: 381 ---LWNPS---INLKSKSSPTIDRF---IYYGFGYDQVNHKYKLLAVK--------AFSR 509
Query: 187 LLKADIYDLSTKSWTPLVSHPPITMVTRIQPSR--YNTLVNGVYYWITSSDGSDAARILC 244
+ + IY S + + R P+R V+G WI + A IL
Sbjct: 510 ITETMIYTFGENS----CKNVEVKDFPRYPPNRKHLGKFVSGTLNWIV-DERDGRATILS 674
Query: 245 FDFRDNQFRKLEAPKLGHYI 264
FD +R++ P+ G+ +
Sbjct: 675 FDIEKETYRQVLLPQHGYAV 734
>TC89815
Length = 807
Score = 67.0 bits (162), Expect = 1e-11
Identities = 51/189 (26%), Positives = 87/189 (45%), Gaps = 12/189 (6%)
Frame = +1
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT-----RRNRLLILQNAPNMKFIFC 83
E+ I+IL LP L RF+ KS +II S F N LI+++ N+ +
Sbjct: 55 EIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLHLKNSNNFYLIIRHNANLYQL-- 228
Query: 84 DGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLI 143
D N IP+ RI ++GSC+G+ C+ I+ D + WNP ++ +I
Sbjct: 229 DFPNLTPPIPLNHPLMSYSNRITLFGSCNGLICISNIA------DDIAFWNPNIRKHRII 390
Query: 144 PRAPSLGNHYSDESL-------YGFGAVNDDFKVVKLNISNSNRMAKINSLLKADIYDLS 196
P P+ SD +L +G+ + D+K+V+++ + +S ++ ++ L
Sbjct: 391 PYLPTTPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDLQNRSFDSQVR--VFSLK 564
Query: 197 TKSWTPLVS 205
SW L S
Sbjct: 565 MNSWKELPS 591
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 66.6 bits (161), Expect = 1e-11
Identities = 52/162 (32%), Positives = 74/162 (45%), Gaps = 17/162 (10%)
Frame = +1
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT---------RRNRLLILQNAP--- 76
EL I+ILL LP KSL RFKCV KSW ++I +FA R L +L + P
Sbjct: 118 ELIIQILLWLPVKSLLRFKCVCKSWFSLISDTHFANSHFQITAKHSRRVLFMLNHVPTTL 297
Query: 77 --NMKFIFCDGGNDQKSIPIKSLFPQDVARIEI-YGSCDGVFCLKGISSCITRHDQLILW 133
+ + + CD + PI + ++ SC G L L +W
Sbjct: 298 SLDFEALHCDNAVSEIPNPIPNFVEPPCDSLDTNSSSCRGFIFLH-------NDPDLFIW 456
Query: 134 NPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKL 173
NP+T+ IP +P+ N + LYGFG + DD+ VV +
Sbjct: 457 NPSTRVYKQIPLSPNDSNSF--HCLYGFGYDQLRDDYLVVSV 576
>TC82343 similar to GP|10177628|dbj|BAB10775.
gb|AAF30317.1~gene_id:K6M13.17~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 65.1 bits (157), Expect = 4e-11
Identities = 53/164 (32%), Positives = 76/164 (46%), Gaps = 16/164 (9%)
Frame = +2
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF---------ATRRNRLLILQNAP 76
L +EL +IL+RLP KSL RFK V KSW ++I +F AT R+L L P
Sbjct: 65 LPDELITKILVRLPVKSLIRFKSVCKSWFSLISDNHFANSHFQVTAATHTLRILFLTATP 244
Query: 77 NMKFI-----FCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLI 131
++I F D N+ + P+ +EI SC G + +C +
Sbjct: 245 EFRYIAVDSLFTDDYNEPVPLNPNFPLPEFEFDLEIKASCRGFIY---VHTC----SEAY 403
Query: 132 LWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKL 173
+WNP+T + IP P++ N YGFG DD+ VV +
Sbjct: 404 IWNPSTGFLKQIPFPPNVSNLI----FYGFGYDESTDDYLVVSV 523
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 64.7 bits (156), Expect = 5e-11
Identities = 62/192 (32%), Positives = 85/192 (43%), Gaps = 17/192 (8%)
Frame = +1
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSW------NNIIKSPY---FATRRNRLLILQN-- 74
L EL I+I+LRLP KSL RFKCV KS +N KS + AT NR++ +
Sbjct: 328 LPHELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNRIVFMSTLA 507
Query: 75 APNMKFIFCDGGNDQK---SIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLI 131
F ND S+ + + P+ + +EI SC G L T +
Sbjct: 508 LETRSIDFEASLNDDSASTSLNLNFMPPESYSSLEIKSSCRGFIVL-------TCSSNIY 666
Query: 132 LWNPTTKEVHLIP-RAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMAKINSLL 188
LWNP+T IP A +L YS LYGFG + DD+ VV ++ + S N
Sbjct: 667 LWNPSTGHHKQIPFPASNLDAKYSC-CLYGFGYDHLRDDYLVVSVSYNTSIDPVDDNISS 843
Query: 189 KADIYDLSTKSW 200
+ L +W
Sbjct: 844 HLKFFSLRANTW 879
>BG647288 weakly similar to GP|5903055|gb|A May be a pseudogene of F6D8.29.
{Arabidopsis thaliana}, partial (12%)
Length = 740
Score = 62.8 bits (151), Expect = 2e-10
Identities = 53/180 (29%), Positives = 84/180 (46%), Gaps = 26/180 (14%)
Frame = +2
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT-------------RRNRLLIL 72
+ +L IL +LP KSL RF CV+KS++ + +SP F + N LLIL
Sbjct: 74 ISNDLAFSILSKLPLKSLKRFTCVKKSFSLLFESPDFMSMFRTNFISKHDENNENTLLIL 253
Query: 73 QNAPNM---KFIFCDGGNDQKSIPIKSLFPQDVAR---IEIYG--SCDGVFCLKGISSCI 124
+ M + FC D+ + FP + + IEI G S +G CL +
Sbjct: 254 KERTQMIPFPYTFCTFAGDKLEDGERLDFPPPLIKGIQIEILGCASVNGTLCLYQGNYGN 433
Query: 125 TRHDQLILWNPTTKEVHLIPRAPSLGNHYSDE-----SLYGFGAVNDDFKVVKLNISNSN 179
T+ ++LWNP T E ++P PS + + E +G+ V D+K++++ SN
Sbjct: 434 TK---IVLWNPATTEFKVVP--PSFQMYDNIELKTRPKAFGYDRVRYDYKLIRIAFYPSN 598
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 62.4 bits (150), Expect = 3e-10
Identities = 54/244 (22%), Positives = 91/244 (37%), Gaps = 20/244 (8%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA-------TRRNRLLILQNAPNMKFI 81
EL EIL RLP K L + +C+ KS+N++I P FA T + L++ N + +F
Sbjct: 183 ELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTPHHLILRSNNGSGRFA 362
Query: 82 FCDGGND-----------QKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQL 130
Q + + ++ A + SCDG+ CL T +
Sbjct: 363 LIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWCSCDGIICL------TTDYSSA 524
Query: 131 ILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMAKINSLL 188
+LWNP + +P + S L+ FG D++KV +
Sbjct: 525 VLWNPFINKFKTLPPLKYISLKRSPSCLFTFGYDPFADNYKVFAITFCVKR--------T 680
Query: 189 KADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFR 248
+++ + T SW + P + + V G +W+T I+ D
Sbjct: 681 TVEVHTMGTSSWRRIEDFPSWSFI-----PDSGIFVAGYVHWLTYDGPGSQREIVSLDLE 845
Query: 249 DNQF 252
D +
Sbjct: 846 DESY 857
>BG646527
Length = 780
Score = 61.2 bits (147), Expect = 6e-10
Identities = 62/215 (28%), Positives = 98/215 (44%), Gaps = 43/215 (20%)
Frame = +1
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT--------RRNRLLILQNAPNMKF 80
++ IL +LP KSL RF CV K W N + P F + N L+L+ P+ F
Sbjct: 25 DISFLILSKLPVKSLKRFTCVCKFWANFFEKPQFMSMYLSKYDDHHNSRLLLKQTPSF-F 201
Query: 81 IFCD--------GGNDQKSIPIKSLFPQDVAR--IEIYGS-CDGVFCL-----KGISSCI 124
+ D G + S+ + P DV R I I GS +G+ CL +G ++ I
Sbjct: 202 DYDDHDYLFLLLGETFENSVKLDWPPPIDVDREGILIAGSIVNGILCLCQGNGRGDTTWI 381
Query: 125 TRHDQLILWNPTTKEVHLIP---------RAPSLGNHYSD-------ESLYGFG--AVND 166
+ +++LWNP+T+E +IP +A G + D +++GFG V+D
Sbjct: 382 AQ--KVVLWNPSTEEFKVIPNGSFEHAILKAFPPGTVFEDLPVINTIVNIHGFGYDPVSD 555
Query: 167 DFKVVKLNISNSN-RMAKINSLLKADIYDLSTKSW 200
D+K+++ N N + IYDL + W
Sbjct: 556 DYKLIRCFCFFDNPEKRDPNDEILWQIYDLRSNCW 660
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 58.2 bits (139), Expect = 5e-09
Identities = 46/197 (23%), Positives = 91/197 (45%), Gaps = 17/197 (8%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGND 88
E+++EIL RLP K L +F+CV K W + I P F + R+ ++ + F
Sbjct: 12 EIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTRHLFLLTFSKLSPELV 191
Query: 89 QKSIPIKSLFPQDVARI--------------EIYGSCDGVFCLKGISSCITRHDQLILWN 134
KS P+ S+F + + GSC G+ C++ S +LWN
Sbjct: 192 IKSYPLSSVFTEMTPTFTQLEYPLNNRDESDSMVGSCHGILCIQCNLSF------PVLWN 353
Query: 135 PTTKEVHLIPRAPSLGNHYSDES-LYGFGAVNDDFKVVKLNISNS--NRMAKINSLLKAD 191
P+ ++ +P N + + + +G+ +D +KVV + +++ N + ++ +L+ +
Sbjct: 354 PSIRKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGVYQLKTLV--N 527
Query: 192 IYDLSTKSWTPLVSHPP 208
++ + T W + + P
Sbjct: 528 VHTMGTNCWRRIQTEFP 578
>BE998730
Length = 820
Score = 57.0 bits (136), Expect = 1e-08
Identities = 48/190 (25%), Positives = 85/190 (44%), Gaps = 17/190 (8%)
Frame = +3
Query: 35 LLRLPTKSLSRFKCVQKSWNNIIKSPYFATRR-----NRLLILQNAPNMKFIFCDGGN-D 88
LL +KSL RF+ KS +II S F NR LI++ ++ I D +
Sbjct: 63 LLPHTSKSLLRFRSTSKSLKSIIDSHNFTNLHLKNSLNRSLIIRRNYDIYQIDDDFSDLT 242
Query: 89 QKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHD---QLILWNPTTKEVHLIPR 145
P+ F + +I + GSC+G+ + +T+ + Q+ +WNP T++ H+IP
Sbjct: 243 TAEFPLNHPFSGNSTKITLIGSCNGLLAISNGLIALTQPNDANQIAIWNPNTRKHHIIPF 422
Query: 146 APSLGNHYSDESLY--------GFGAVNDDFKVVKLNISNSNRMAKINSLLKADIYDLST 197
P + +Y GF + D+K+++++ + +S ++ L T
Sbjct: 423 LPLPITPHPPPDMYCYLYLHGFGFDLLTADYKLLRISWLVTQHNPFYDS--HVTLFSLKT 596
Query: 198 KSWTPLVSHP 207
SW + S P
Sbjct: 597 NSWKTIPSTP 626
>BG648114 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.17~similar
to unknown protein {Arabidopsis thaliana}, partial (5%)
Length = 734
Score = 55.5 bits (132), Expect = 3e-08
Identities = 36/131 (27%), Positives = 64/131 (48%), Gaps = 19/131 (14%)
Frame = +1
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAP---------NM 78
+++ IL +LP KSL RF+CV+KSW+ + ++PYF R+ + +N +
Sbjct: 274 DDVAFVILSKLPLKSLFRFRCVRKSWSLLFENPYFMDMFRRIFLSKNHSYYNDTSLLLHD 453
Query: 79 KFIFCDGGNDQKSIPIKSLFPQDVAR---IEIYG--SCDGVFCLKGISS-----CITRHD 128
+++ ++ +K +P IYG S +G+ C++ CI
Sbjct: 454 EYMLYSLFGERFENRVKLDWPNPYGEQFDFNIYGCASVNGILCIEDAGRIEGIHCIEELG 633
Query: 129 QLILWNPTTKE 139
+++LWNPTT E
Sbjct: 634 RVVLWNPTTGE 666
>TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pseudogene of
F6D8.29. {Arabidopsis thaliana}, partial (10%)
Length = 1210
Score = 54.7 bits (130), Expect = 6e-08
Identities = 45/177 (25%), Positives = 77/177 (43%), Gaps = 31/177 (17%)
Frame = +1
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT--RRNRLLILQNA---------- 75
++L IL +LP KSL+RF C QKSW+ + ++ F R N LL +
Sbjct: 94 DDLTFSILSKLPLKSLTRFTCAQKSWSLLFQNSIFMNMFRTNFLLSKHDEDDENTCVLLK 273
Query: 76 ------PNMKFIFCDGGNDQKS-----IPIKSLFPQDVARIEIYGSCDGVFCLKGISSCI 124
P ++ G ++ +P L RI +GV CL S +
Sbjct: 274 QMEGRRPYHTSLYTLSGEKFENSVRLDLPTPLLENDSFIRILCSAGINGVLCLYNDS--L 447
Query: 125 TRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESL--------YGFGAVNDDFKVVKL 173
+ ++LWNP T+E ++P +H E++ +G+ V +D+KV+++
Sbjct: 448 NANKTIVLWNPATEEFKVVPH-----SHQPYENIEFNPRPFAFGYDPVTNDYKVIRV 603
>TC83174
Length = 1323
Score = 54.7 bits (130), Expect = 6e-08
Identities = 52/201 (25%), Positives = 91/201 (44%), Gaps = 18/201 (8%)
Frame = +3
Query: 130 LILWNPTTKEVHLIPRAPSLGNHYSDESL-------YGFGAVNDDFKVVKLNISNSNRMA 182
L++ NP T E +P A + + + + +GF +++KV+K+ I + R
Sbjct: 699 LVICNPVTGEFIRLPEATTTPLRLNTDRVRMQGQAGFGFQPKTNEYKVIKIWIRHVQRAN 878
Query: 183 K-INSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAAR 241
+ + +I L SW + P I+ I +Y T VNGV +WI +G +
Sbjct: 879 DWVFDRVILEINTLGAPSWRTVEVDPRIS----ISSLKYPTCVNGVLHWI-KFEGQQKS- 1040
Query: 242 ILCFDFRDNQFRKLEAPKLGHYIPFFCD---DVFEIKGYLGYVVQYRCRI---VWLEIWT 295
ILCF F + + +P H + + E+KG+L Y C + V + +W
Sbjct: 1041ILCFCFEGERLQPFPSPP--HVFGIHDNRRISMGELKGFL-----YICDLNFFVDVTMWV 1199
Query: 296 LEQNG----WAKKYNIDTKMS 312
+ ++G W K YNI+T ++
Sbjct: 1200MNEHGIEESWTKVYNIETSVN 1262
>TC89054 weakly similar to GP|5903055|gb|AAD55614.1| May be a pseudogene of
F6D8.29. {Arabidopsis thaliana}, partial (13%)
Length = 825
Score = 54.3 bits (129), Expect = 7e-08
Identities = 48/172 (27%), Positives = 78/172 (44%), Gaps = 27/172 (15%)
Frame = +1
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT-------------RRNRLLILQNA 75
+L IL +LP KSL RF CV+KS++ + +SP F N LIL+
Sbjct: 82 DLAFSILSKLPLKSLKRFTCVKKSFSLLFQSPDFMNMFRTNFISKHNEDNENIRLILKEK 261
Query: 76 PNM---KFIFC----DGGNDQKSIPIKSLFPQDVARIEIYG--SCDGVFCLKGISSCITR 126
M + C + D + + F +D IEI+G S +G CL I
Sbjct: 262 TQMIPFPYTLCTFSGEKLEDGERLDCPPPFHEDDLGIEIFGFASVNGTLCL---YQGIYY 432
Query: 127 HDQLILWNPTTKEVHLIPRAPSLGNHYSDE-----SLYGFGAVNDDFKVVKL 173
+++LWNP T + ++P PS + + E +G+ V D+K++++
Sbjct: 433 DTKIVLWNPATTKFKVVP--PSFQPYDNIEVKTPPLAFGYDHVRHDYKLIRI 582
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,539,626
Number of Sequences: 36976
Number of extensions: 234533
Number of successful extensions: 1199
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 1162
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1175
length of query: 373
length of database: 9,014,727
effective HSP length: 98
effective length of query: 275
effective length of database: 5,391,079
effective search space: 1482546725
effective search space used: 1482546725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135160.10


