

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135101.10 + phase: 0 /pseudo
(1477 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
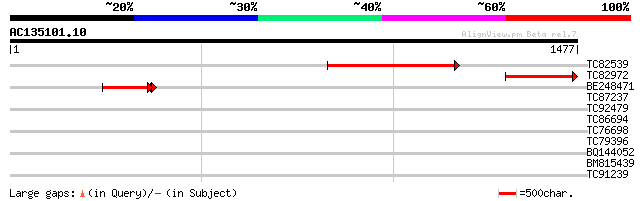
Score E
Sequences producing significant alignments: (bits) Value
TC82539 686 0.0
TC82972 weakly similar to PIR|T05672|T05672 hypothetical protein... 358 1e-98
BE248471 similar to PIR|T05673|T05 hypothetical protein F20M13.1... 250 2e-68
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 35 0.17
TC92479 similar to GP|9759160|dbj|BAB09716.1 contains similarity... 34 0.37
TC86694 similar to PIR|T05949|T05949 phosphatidylinositol-phosph... 32 1.8
TC76698 similar to PIR|S50766|S50766 dehydrin-related protein - ... 30 5.3
TC79396 similar to GP|13605881|gb|AAK32926.1 At2g11520/F14P14.15... 30 7.0
BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU09387... 30 7.0
BM815439 30 7.0
TC91239 similar to GP|6598622|gb|AAF18655.1| unknown protein {Ar... 30 9.1
>TC82539
Length = 1032
Score = 686 bits (1771), Expect = 0.0
Identities = 339/343 (98%), Positives = 341/343 (98%)
Frame = +2
Query: 829 VTKSGRDSGDESFMKELIHLRQKGDIEMSLASTCCLNGIINVITKIDNLIRSAKTGICNP 888
VTKSGRDSGDESFMKELIHLRQKGDIEMSLASTCCLNGIINVITKIDNLIRSAKTGICNP
Sbjct: 2 VTKSGRDSGDESFMKELIHLRQKGDIEMSLASTCCLNGIINVITKIDNLIRSAKTGICNP 181
Query: 889 PVTEQSLSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAPGMG 948
P TEQS SKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAPGMG
Sbjct: 182 PGTEQSQSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAPGMG 361
Query: 949 VGWGAHGGGFWSKTVLPVKTDARLLVCLLQIFENTSNDAPETEQMTFSMQQVNTALGLCL 1008
VGWGAHGGGFWSKTVLPV+TDARLLVCLLQIFENTSNDAPETEQMTFSMQ+VNTALGLCL
Sbjct: 362 VGWGAHGGGFWSKTVLPVQTDARLLVCLLQIFENTSNDAPETEQMTFSMQRVNTALGLCL 541
Query: 1009 TAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMHFSRMLSS 1068
TAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMHFSRMLSS
Sbjct: 542 TAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMHFSRMLSS 721
Query: 1069 HFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSDMSSTTSPCCNSLMIEWAR 1128
HFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSDMSSTTSPCCNSLMIEWAR
Sbjct: 722 HFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSDMSSTTSPCCNSLMIEWAR 901
Query: 1129 QNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANLLEV 1171
QNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANLLEV
Sbjct: 902 QNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANLLEV 1030
>TC82972 weakly similar to PIR|T05672|T05672 hypothetical protein F22I13.210 -
Arabidopsis thaliana, partial (6%)
Length = 569
Score = 358 bits (918), Expect = 1e-98
Identities = 180/185 (97%), Positives = 181/185 (97%)
Frame = +2
Query: 1293 FGRQVSVYLHCCVESSIRLATWNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEA 1352
FGRQVSVYLH CVESSIRLATWNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEA
Sbjct: 2 FGRQVSVYLHRCVESSIRLATWNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEA 181
Query: 1353 YAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACPVDKLLLRNNLVRSLLRDYAGKQQ 1412
YAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACPVDKLLLRN LVRSLLRDYAGKQQ
Sbjct: 182 YAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACPVDKLLLRNRLVRSLLRDYAGKQQ 361
Query: 1413 HEGMLMNLISHNRQSTSNMDEQLDGLLHEESWLESRMKVLIEACEGNSSLLIQVKKLKDA 1472
HEGMLMNLISHN+QSTSNMDEQLDGLLHEESWLESRMKVL EACEGNSSLL QVKKLKDA
Sbjct: 362 HEGMLMNLISHNKQSTSNMDEQLDGLLHEESWLESRMKVLNEACEGNSSLLTQVKKLKDA 541
Query: 1473 AEKNS 1477
AEKNS
Sbjct: 542 AEKNS 556
>BE248471 similar to PIR|T05673|T05 hypothetical protein F20M13.10 -
Arabidopsis thaliana, partial (4%)
Length = 444
Score = 250 bits (638), Expect(2) = 2e-68
Identities = 128/134 (95%), Positives = 128/134 (95%), Gaps = 4/134 (2%)
Frame = +2
Query: 242 EDDISHTIMAPPSKKQLDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDVVDTE 301
EDDISHTIMAPPSKKQLDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDVVDTE
Sbjct: 2 EDDISHTIMAPPSKKQLDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDVVDTE 181
Query: 302 QEPVYDNIAERDYLRTEGDPGAAGYTIKEALEITRSV----RALGLHLLSSVLDKALCYI 357
QEPVYDNIAERDYLRTEGDPGAAGYTIKEALEITRSV RALGLHLLSSVLDKALC I
Sbjct: 182 QEPVYDNIAERDYLRTEGDPGAAGYTIKEALEITRSVIPGQRALGLHLLSSVLDKALCCI 361
Query: 358 CKDRTENMTKKGNK 371
CKDRT NMTKKGNK
Sbjct: 362 CKDRTGNMTKKGNK 403
Score = 29.3 bits (64), Expect(2) = 2e-68
Identities = 13/17 (76%), Positives = 14/17 (81%), Gaps = 1/17 (5%)
Frame = +1
Query: 367 KKGN-KVDKSVDWEAVW 382
KKG + DKSVDWEAVW
Sbjct: 388 KKGEQRSDKSVDWEAVW 438
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 35.4 bits (80), Expect = 0.17
Identities = 61/303 (20%), Positives = 117/303 (38%), Gaps = 17/303 (5%)
Frame = +1
Query: 40 REFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGGKNTSKKISSYSDGSVFASMEVDAKP 99
++ T++ G + E+ T+ + ++G KN + +YS G +S VD K
Sbjct: 466 KQGTEETNGTEGGEKEEHDKIKEDTSSDNQVQDGEKNNEAREENYS-GDNASSAVVDNKS 642
Query: 100 QLV--KLDGGFI-------------NSATSMELDTSNKDDKKEVFAAERDKIFSDRMTDH 144
Q K + F NS + E S + +E+D+ ++ T
Sbjct: 643 QESSNKTEEQFDKKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPK 822
Query: 145 SSTSEKNYFMHEQE--STSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQ 202
S SE + EQE +T+ ++ ++ ++ +TE+ E D K
Sbjct: 823 GSASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDTTEKQNETSEDANSK---------- 972
Query: 203 KRGKEKLKKPNSLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKN 262
KE N+ + + + Q T +T+D ++ + + +KN
Sbjct: 973 ---KEDSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTNENPEKN 1143
Query: 263 VSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDVVDTEQEPVYDNIAERDYLRTEGDPG 322
SG+ T S SS N + N+ A +++ + D +EQ+ + AE ++ D
Sbjct: 1144-SGQEGTQESGSSSNTFDNKDAASNKVQLTTTSD-TSSEQKKDESSSAESKSESSQNDNA 1317
Query: 323 AAG 325
+G
Sbjct: 1318NSG 1326
>TC92479 similar to GP|9759160|dbj|BAB09716.1 contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (4%)
Length = 844
Score = 34.3 bits (77), Expect = 0.37
Identities = 42/175 (24%), Positives = 79/175 (45%), Gaps = 6/175 (3%)
Frame = +2
Query: 64 TTGRKKNENGGKNT--SKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSA-TSMELDTS 120
+ GR + +G K +KKI+S S S E+ + + +K+ G I S T++E T+
Sbjct: 95 SNGRNQLSDGSKTIPINKKIAS----SPHHSKEMASLREKIKMLEGLIKSKETALETSTT 262
Query: 121 NKDDKKEVFAAERDKIFSDRMTDHSSTSEK---NYFMHEQESTSLENEIDSENRARIQQM 177
+ + +++K R+ + + E+ N +HE S NEI + R R+
Sbjct: 263 S--------SMKKEKELQSRIVELENKVEEFNQNVTLHEDRSIKSSNEISEKVRNRL--- 409
Query: 178 STEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQQVQI 232
E + + + ++ ++S LK K + +LK+ SE+ V + QI
Sbjct: 410 --EHADNSLSGVLTELSS--LKERNKSMESELKEMQERYSEMSLKFAEVEGERQI 562
>TC86694 similar to PIR|T05949|T05949 phosphatidylinositol-phosphatidylcholine
transfer protein SEC14 homolog Ssh1 - soybean, partial
(98%)
Length = 1433
Score = 32.0 bits (71), Expect = 1.8
Identities = 19/62 (30%), Positives = 35/62 (55%)
Frame = +3
Query: 1233 QDLYGELLDKERFNQNKEAISDDKKHIEFLRFKSDIHESYSTFIEELVEQFSSISYGDLI 1292
Q+LY + ++ R N++++ I H+EF +D E T IE + +F + S+G++I
Sbjct: 981 QELYNYIKEQSRMNEDRKPIKHGSFHVEFPEPSADDGEIAKT-IESEIHKFEN-SHGNVI 1154
Query: 1293 FG 1294
G
Sbjct: 1155 DG 1160
>TC76698 similar to PIR|S50766|S50766 dehydrin-related protein - garden pea,
partial (80%)
Length = 974
Score = 30.4 bits (67), Expect = 5.3
Identities = 35/147 (23%), Positives = 60/147 (40%), Gaps = 5/147 (3%)
Frame = +2
Query: 77 TSKKISSYSDGSVFASM-----EVDAKPQLVKLDGGFINSATSMELDTSNKDDKKEVFAA 131
T+ + + D VF + + + KPQ + F + T E + K ++KE
Sbjct: 128 TNSETTEIKDRGVFDFLGGKKKDEEHKPQEEAIATDFNHKVTLYEAPSETKVEEKE---- 295
Query: 132 ERDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIME 191
E +K H+S EK + S+S E E+D E R + ++ E+ A +E
Sbjct: 296 EGEK-------KHTSLLEKLHRSDSSSSSSSEEEVDGEKRKKKKKEKKEDTSVA----VE 442
Query: 192 KISPALLKVLQKRGKEKLKKPNSLKSE 218
K+ + K K K P K++
Sbjct: 443 KVDGTTEEKKGFLDKIKEKLPGHKKTD 523
>TC79396 similar to GP|13605881|gb|AAK32926.1 At2g11520/F14P14.15 {Arabidopsis
thaliana}, partial (53%)
Length = 1658
Score = 30.0 bits (66), Expect = 7.0
Identities = 18/51 (35%), Positives = 24/51 (46%)
Frame = +1
Query: 403 NHNSVVLACAKVVQSALSCDVNENYFDISENMATYDKDICTAPVFRSRPDI 453
N SVV ++Q A+ DV FD++ N C APV RPD+
Sbjct: 1000 NEGSVVALLDPLMQEAVKTDVAVKMFDLAFN--------CAAPVRSDRPDM 1128
>BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU093876(E1018)
AU093877(E1018)~unknown protein, partial (11%)
Length = 536
Score = 30.0 bits (66), Expect = 7.0
Identities = 15/26 (57%), Positives = 15/26 (57%), Gaps = 3/26 (11%)
Frame = -1
Query: 937 IFGRGGPAPGMGVG---WGAHGGGFW 959
I GRGG A G G G WG GGG W
Sbjct: 347 IAGRGGGAMGNGKGNGGWGRKGGGNW 270
>BM815439
Length = 705
Score = 30.0 bits (66), Expect = 7.0
Identities = 43/216 (19%), Positives = 83/216 (37%), Gaps = 6/216 (2%)
Frame = +1
Query: 40 REFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGGKNTSKKISSYSDGSVFASMEVDA-- 97
RE DK+ +G++ + V K ++G + T K S G + + +
Sbjct: 136 RERKNKDKNLVGREKPEVVDV---VVEEKNVDHGMQVTEGKKVKVSTGRKRKNKDKNLVG 306
Query: 98 --KPQLVKLDGGFINSATSMELDTSNKDDKKEVFAAERDKIFSDRMTDHSSTSEKNYFMH 155
KP+ V++D L+ N D + E+ + K + R + +KN
Sbjct: 307 REKPETVEVD-----------LEEQNVDPEMEIIKGRKVKATTSRKRKNK---DKNLVGR 444
Query: 156 EQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPNSL 215
+ ++ E ++ EE++++K + A+ K+ K+K +K
Sbjct: 445 QTPEDEVDFEEKNDGVVEHSDSKEEEVKDSKKHRDSNVEAAIKPCKSKQAKKKRRK---- 612
Query: 216 KSEVGAVTESVNQQVQITQGAKHL--QTEDDISHTI 249
V +S ++ Q Q H+ +DD SH I
Sbjct: 613 -----EVIKSPEKREQNHQDDIHIISSGDDDCSHGI 705
>TC91239 similar to GP|6598622|gb|AAF18655.1| unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 743
Score = 29.6 bits (65), Expect = 9.1
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = +3
Query: 647 YCVSHFSKIFPALCFWLDLPSFEKLTKNNVLNES 680
+C+ S I LC+ L L +F K +K +L ES
Sbjct: 243 HCILGHSSICTTLCYMLSLGTFPKFSKVRILRES 344
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,181,953
Number of Sequences: 36976
Number of extensions: 658320
Number of successful extensions: 3520
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 3457
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3517
length of query: 1477
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1369
effective length of database: 5,021,319
effective search space: 6874185711
effective search space used: 6874185711
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC135101.10


