

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135100.7 + phase: 0 /pseudo
(855 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
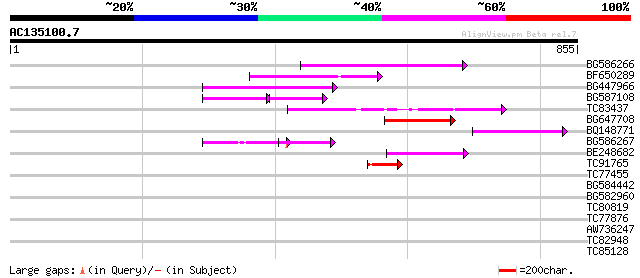
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 170 2e-42
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 139 6e-33
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 133 3e-31
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 70 5e-25
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 97 2e-20
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 88 2e-17
BQ148771 81 2e-15
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 62 9e-10
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 57 2e-08
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 49 8e-06
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 42 0.001
BG584442 38 0.019
BG582960 33 0.36
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 33 0.36
TC77876 similar to PIR|T05265|T05265 coat protein gamma-COP homo... 31 1.8
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 30 3.0
TC82948 30 3.0
TC85128 similar to PIR|D84577|D84577 probable recA protein [impo... 29 8.8
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 170 bits (431), Expect = 2e-42
Identities = 88/253 (34%), Positives = 145/253 (56%), Gaps = 1/253 (0%)
Frame = -3
Query: 439 LKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRGTTDNAIIAQEVVHYM 498
+ +R I+ CN +K+I K+L R++P L ++SP QS F+P R +DN +I +++HY+
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 499 HTSRSQKG-TLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMSCVKASDLAILWN 557
S ++K ++A+K + KAYDR+ W+FL L GF I IM CV + L N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 558 GAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLT 617
G + P++GLRQGDPLSPYLF+LC E L+ Q + + + ++++ P ++HL
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLL 239
Query: 618 FADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISS 677
FADD + F ++N +++ ++++ +ASG +N TKS S + + ++G
Sbjct: 238 FADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELK 59
Query: 678 IRLVSNLGHYLGF 690
I G YLG+
Sbjct: 58 IAKEGGTGKYLGY 20
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 139 bits (349), Expect = 6e-33
Identities = 80/203 (39%), Positives = 120/203 (58%), Gaps = 3/203 (1%)
Frame = +3
Query: 362 LSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEA 421
L S EVK+ALFSM S KAPG DG+ FFK W G+S+ + + F+ GF+ +
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 422 IVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPR 481
I T + L+PKE N T +KNFRPI+ C+VI+K+I+K+L +R++ L+ +VS QS F+
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 482 RGTTDNAIIAQEVVHYMHTSRSQKG---TLALKIYLEKAYDRLDWSFLELTLHDFGFPHH 538
R DN I++ E+V S S+KG +KI L KAYD +W F++ + + GFP+
Sbjct: 372 RVIFDNIILSHELV----KSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 539 IIGLIMSCVKASDLAILWNGAKT 561
+ +M+ + + NG T
Sbjct: 540 FVNWVMAXLTTASYTFNXNGDLT 608
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 133 bits (334), Expect = 3e-31
Identities = 73/203 (35%), Positives = 109/203 (52%)
Frame = +2
Query: 292 FFHTQTIVRRKHNKIHGLHLDDGTWCTYDTQLQKTAISFYQNLFQKDRTVNPHSLRVPLI 351
FFH++ RRK N+I L + G WC + +++ I+++ NLF +
Sbjct: 35 FFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPTAIEETCEVVK 214
Query: 352 PTLCALGIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDI 411
L I +EEV +A+ M KAPGPDG LFF+ +W G+ + ++V
Sbjct: 215 GKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIVGKEVQQMVLQ 394
Query: 412 SFRRGFIDEAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELV 471
E + +T +VLIPK NP K++RPISLCNV+ K+ITKV+ NR++ L +++
Sbjct: 395 VLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQTLPDVI 574
Query: 472 SPFQSNFIPRRGTTDNAIIAQEV 494
QS F+ R TDNA+IA V
Sbjct: 575 DVEQSAFVQGRLITDNALIAWSV 643
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 69.7 bits (169), Expect(2) = 5e-25
Identities = 35/87 (40%), Positives = 51/87 (58%)
Frame = +3
Query: 393 FFKHFWEKTGESLWELVDISFRRGFIDEAIVETLLVLIPKENNPTHLKNFRPISLCNVIF 452
FF+H W L ++V+ G +D + T + LIPK+ PT + RPISLCNV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 453 KVITKVLVNRIRPFLDELVSPFQSNFI 479
K+I+KVL R++ L L+S QS F+
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFV 671
Score = 63.9 bits (154), Expect(2) = 5e-25
Identities = 34/103 (33%), Positives = 53/103 (51%)
Frame = +2
Query: 292 FFHTQTIVRRKHNKIHGLHLDDGTWCTYDTQLQKTAISFYQNLFQKDRTVNPHSLRVPLI 351
F+H T R N+I GL+ DG W T + ++K A+ ++++LFQ+ +
Sbjct: 107 FYHALTKQRHARNRIVGLYDYDGNWITEEQGVEKVAVDYFEDLFQRTTPTGFDGFLDEIT 286
Query: 352 PTLCALGIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFF 394
++ + L +EEV+ ALF M +KAPGPDG L F
Sbjct: 287 SSITPQMNQRLLRLATEEEVRLALFIMHPEKAPGPDGMTTLLF 415
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 97.4 bits (241), Expect = 2e-20
Identities = 91/335 (27%), Positives = 147/335 (43%), Gaps = 5/335 (1%)
Frame = +2
Query: 420 EAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFI 479
+ I T + LIPK +NP L +FRPISL ++K++ K+L NR+R + ++S QS F+
Sbjct: 44 KGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFV 223
Query: 480 PRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDR---LDWSFLELTLHDFGFP 536
R + + Q + +M +K L+ L++ L W L G
Sbjct: 224 KNRQILEMVFL*QMRL-WMRLRN*RKIFCCLRWILKRLITLSIGLIW-----ILF*VGMS 385
Query: 537 HHIIGL--IMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQG 594
++ I CV + ++L NG+ TN K L Q + Y F +
Sbjct: 386 FLVLWRKWIKECVSTATTSVLVNGSPTNVLM--KSLVQTQLFTRYSFGVV---------- 529
Query: 595 KVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVT 654
P+ +S HL FA+D LL N ++ + L F + SGLKVN
Sbjct: 530 -------NPVVVS-------HLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFH 667
Query: 655 KSKSMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHHRLA 714
KS +C P + + S ++ YLG P+ ++ I+ ++ RL
Sbjct: 668 KSGLVCVNIAPSWLS-EAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKARLT 844
Query: 715 SWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRK 749
W + L+ GR+ L KSV TS+ VY++ + L +
Sbjct: 845 GWNSRFLSFGGRLVLLKSVLTSLSVYALPSSKLHQ 949
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 87.8 bits (216), Expect = 2e-17
Identities = 45/107 (42%), Positives = 65/107 (60%)
Frame = +1
Query: 566 PTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLF 625
P KGLRQGDPLSPYLF+LC L+ ++ + ++ I +++ P ++HL FADD LLF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 626 CEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDI 672
AN + ++ L+ + SASG VN KS+ S+ VP + K I
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMI 324
>BQ148771
Length = 680
Score = 80.9 bits (198), Expect = 2e-15
Identities = 45/144 (31%), Positives = 76/144 (52%)
Frame = -3
Query: 698 TNETYKHIIEKMHHRLASWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGIDK 757
T ++ + ++H LA+WK L+ + RV L KSV ++P+Y M ++ K + I K
Sbjct: 618 TKKSRFSVYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQK 439
Query: 758 ITKSFIWGGNGVT*KWNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDKNKL 817
+ + F+WG V+ +++ V W T++ P+ GL +R N A + KL WS+ N L
Sbjct: 438 LQRKFVWGDTEVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSL 259
Query: 818 WVRLLSGKIY*EWLFMEQRSSEKP 841
++ GK Y +E+ EKP
Sbjct: 258 CTEVMRGK-YQRSESLEEIFLEKP 190
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 62.0 bits (149), Expect = 9e-10
Identities = 42/137 (30%), Positives = 66/137 (47%), Gaps = 3/137 (2%)
Frame = +3
Query: 292 FFHTQTIVRRKHNKIHGLHLDDGTWCTYDTQLQKTAISFYQNLFQKDR---TVNPHSLRV 348
FFH T RR N+I L DD + L + A S ++ L+ + T+ + +
Sbjct: 189 FFHAVTKNRRAQNRILSLIDDDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWN-SI 365
Query: 349 PLIPTLCALGIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWEL 408
P I T L + + +EEV++A+F + K PGPDG FF+ FW+ G+ L +
Sbjct: 366 PAIVT--EEQNAQLMAQISREEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSM 539
Query: 409 VDISFRRGFIDEAIVET 425
R G ++E I +T
Sbjct: 540 AQEFLRTGKLEEGINKT 590
Score = 55.1 bits (131), Expect = 1e-07
Identities = 30/91 (32%), Positives = 51/91 (55%), Gaps = 5/91 (5%)
Frame = +1
Query: 406 WELVDISFRR-----GFIDEAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLV 460
WE++ +R+ G + + L+PK+ L FRPISLCNV +K+++KVL
Sbjct: 517 WEMISHLWRKNSSEQGNLKRESTKQTSGLVPKKLEAKRLVEFRPISLCNVAYKIVSKVLS 696
Query: 461 NRIRPFLDELVSPFQSNFIPRRGTTDNAIIA 491
R++ L +++ Q+ F R+ +DN +IA
Sbjct: 697 KRLKSVLPWIITETQAAFGRRQLISDNILIA 789
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 57.4 bits (137), Expect = 2e-08
Identities = 41/124 (33%), Positives = 59/124 (47%)
Frame = +3
Query: 568 KGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCE 627
+GL+QGDPL+P+LF+L E ++ ++ V + + + G +SHL +ADD L
Sbjct: 24 RGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGM 203
Query: 628 ANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGHY 687
+ + L F ASGLKVN KS S+ VP + R S Y
Sbjct: 204 PTVDNLWTLKALLQGFEMASGLKVNFHKS-SLIGINVPRDFMEAACRFLNCREESIPFIY 380
Query: 688 LGFP 691
LG P
Sbjct: 381 LGLP 392
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 48.9 bits (115), Expect = 8e-06
Identities = 23/53 (43%), Positives = 36/53 (67%)
Frame = +2
Query: 540 IGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHI 592
IG M CV+++D +L N + P++GL+QGD LSPY+F++C+E L+ I
Sbjct: 89 IGCFM-CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLI 244
Score = 34.7 bits (78), Expect = 0.16
Identities = 19/59 (32%), Positives = 29/59 (48%)
Frame = +3
Query: 647 SGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHI 705
SG +++ KS CS+ VP+ K I I ++ + YLG P + GR T+ I
Sbjct: 372 SGKAISLRKS*IYCSRNVPDILKTSITYILGVQFMLGTCKYLGLPSMIGRDRTTTFSSI 548
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 42.0 bits (97), Expect = 0.001
Identities = 19/39 (48%), Positives = 25/39 (63%)
Frame = -3
Query: 784 PRKFGGLAIRDARNTNLALLGKLVWSLLHDKNKLWVRLL 822
PR GGL +RD R N++LL K W LL D++ LW +L
Sbjct: 963 PRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVL 847
>BG584442
Length = 775
Score = 37.7 bits (86), Expect = 0.019
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 3/110 (2%)
Frame = +1
Query: 717 KGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*KWNMV 776
+ K L+K + K S+ Y M I LL D I+KI +F W G K +
Sbjct: 397 RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGENRKG--M 570
Query: 777 KWTT---VTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDKNKLWVRLLS 823
W + + + +GG+ D N+ +LGK V S L ++ L++ +S
Sbjct: 571 HWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLFLEKIS 720
>BG582960
Length = 852
Score = 33.5 bits (75), Expect = 0.36
Identities = 25/69 (36%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Frame = +3
Query: 485 TDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIG--- 541
T+N +I +E+ + +G LALKI + KA D L FL L F F H
Sbjct: 39 TNNKLILEEINFL--DKKILRGNLALKITMRKALDNLV*EFLLNVLKPFVFKHIFYN*NK 212
Query: 542 LIMSCVKAS 550
I++CVK S
Sbjct: 213 SILNCVKLS 239
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 33.5 bits (75), Expect = 0.36
Identities = 15/27 (55%), Positives = 18/27 (66%)
Frame = +1
Query: 799 NLALLGKLVWSLLHDKNKLWVRLLSGK 825
NL+LLGK W LL DK LW R+L +
Sbjct: 52 NLSLLGKWCWRLLVDKEGLWHRVLKAR 132
>TC77876 similar to PIR|T05265|T05265 coat protein gamma-COP homolog
T4L20.30 - Arabidopsis thaliana, partial (50%)
Length = 1746
Score = 31.2 bits (69), Expect = 1.8
Identities = 15/50 (30%), Positives = 31/50 (62%)
Frame = +1
Query: 427 LVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQS 476
L+L P+++NP HL+ +P+ +++ V+ +V+ ++ + E PFQS
Sbjct: 493 LILCPRKSNPNHLQRRKPLVKSQLVW-VLLQVVPHQQLMHIRERFPPFQS 639
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 30.4 bits (67), Expect = 3.0
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Frame = +1
Query: 368 KEEVKDALFSMQSDKA--PGPDGFQPLFFKHFWE 399
+EEV+ A++ S + PGPDG F K +WE
Sbjct: 127 EEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWE 228
>TC82948
Length = 705
Score = 30.4 bits (67), Expect = 3.0
Identities = 16/41 (39%), Positives = 19/41 (46%)
Frame = +3
Query: 776 VKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDKNK 816
V W V P K G L IR N AL KL W ++ K +
Sbjct: 267 VSWEKVCRPIKEGSLGIRSLSKLNEALNLKLCWDMMISKEQ 389
>TC85128 similar to PIR|D84577|D84577 probable recA protein [imported] -
Arabidopsis thaliana, partial (71%)
Length = 821
Score = 28.9 bits (63), Expect = 8.8
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = +3
Query: 402 GESLWELVDISFRRGFIDEAIVETLLVLIPK 432
GE LVD R G ID +V+++ L+PK
Sbjct: 273 GEQALSLVDTLIRSGSIDVIVVDSVAALVPK 365
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.345 0.152 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,943,065
Number of Sequences: 36976
Number of extensions: 405617
Number of successful extensions: 3594
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1510
Number of HSP's successfully gapped in prelim test: 169
Number of HSP's that attempted gapping in prelim test: 2041
Number of HSP's gapped (non-prelim): 1764
length of query: 855
length of database: 9,014,727
effective HSP length: 104
effective length of query: 751
effective length of database: 5,169,223
effective search space: 3882086473
effective search space used: 3882086473
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135100.7


