

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.4 - phase: 0
(290 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
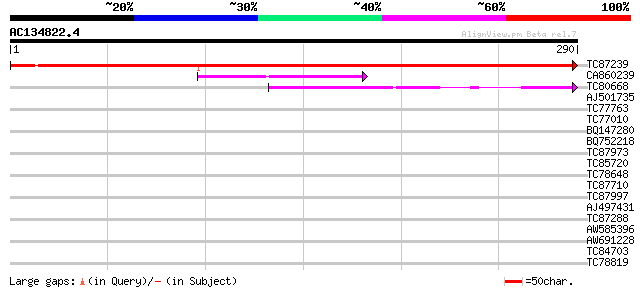
TC87239 similar to PIR|T09028|T09028 hypothetical protein T27E11... 490 e-139
CA860239 weakly similar to SP|P17624|NUDC_ Nuclear movement prot... 71 4e-13
TC80668 similar to GP|18389274|gb|AAL67080.1 unknown protein {Ar... 71 4e-13
AJ501735 similar to GP|18389274|gb unknown protein {Arabidopsis ... 37 0.009
TC77763 similar to GP|15215674|gb|AAK91382.1 AT4g27500/F27G19_10... 34 0.075
TC77010 similar to PIR|S59560|S59560 histone H1.41 - garden pea,... 30 0.83
BQ147280 similar to PIR|C86333|C863 hypothetical protein AAF7991... 30 1.1
BQ752218 similar to GP|19170914|emb hypothetical protein {Enceph... 29 2.4
TC87973 weakly similar to PIR|D86282|D86282 protein F10B6.26 [im... 29 2.4
TC85720 similar to SP|P08283|H1_PEA Histone H1. [Garden pea] {Pi... 28 3.2
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 28 3.2
TC87710 similar to PIR|T08953|T08953 Cdk-activating protein kina... 28 4.1
TC87997 similar to GP|12597886|gb|AAG60194.1 putative phosphatid... 28 5.4
AJ497431 similar to GP|9759308|dbj contains similarity to transf... 27 7.0
TC87288 similar to PIR|S64423|S64423 probable membrane protein Y... 27 7.0
AW585396 27 7.0
AW691228 similar to PIR|H96592|H96 probable multispanning membra... 27 7.0
TC84703 weakly similar to GP|20268746|gb|AAM14076.1 unknown prot... 27 9.2
TC78819 similar to GP|9558421|dbj|BAB03357.1 ESTs AU097577(C6200... 27 9.2
>TC87239 similar to PIR|T09028|T09028 hypothetical protein T27E11.130 -
Arabidopsis thaliana, partial (69%)
Length = 1228
Score = 490 bits (1261), Expect = e-139
Identities = 251/296 (84%), Positives = 263/296 (88%), Gaps = 6/296 (2%)
Frame = +1
Query: 1 MAIISDYQDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDS 60
MAIISDYQ+ETQT SSSSQPKPSK IPFSSTFDPS PTAFLE VFDFIAKESTDFFD DS
Sbjct: 61 MAIISDYQEETQT-SSSSQPKPSKPIPFSSTFDPSNPTAFLENVFDFIAKESTDFFDNDS 237
Query: 61 AEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKA------AAGIKVNDEKSGVGTEKKD 114
AEK+VLSAVRA KVKKAKAVAAEKAKIA++EKAK AA K E +KKD
Sbjct: 238 AEKVVLSAVRAVKVKKAKAVAAEKAKIAAEEKAKVDKAASVAAAEKKAKEVDEKEDDKKD 417
Query: 115 GESGLAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQP 174
GESGLAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKS FV CEIKKNHLKVG+KGQP
Sbjct: 418 GESGLAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSRFVTCEIKKNHLKVGIKGQP 597
Query: 175 PIIDGELYKFIKADECYWSIEDQSTVSILLTKHDQMDWWKCLVKGDPLINTQKVEPESSK 234
PIIDGELYK IK D+CYWSIEDQ+T+SILLTKHDQMDWWKCLVKGDP INTQKVEPESSK
Sbjct: 598 PIIDGELYKSIKPDDCYWSIEDQNTLSILLTKHDQMDWWKCLVKGDPEINTQKVEPESSK 777
Query: 235 LGELDSETRMTVEKMMFDQRQKSMGLPTSEELEKQEMMKKFMSQHPNMDFSGAKLS 290
LG+LD ETR TVEKMMFDQRQKSMGLPTSEEL+KQE+MKKFMSQHP MDFS AKLS
Sbjct: 778 LGDLDPETRQTVEKMMFDQRQKSMGLPTSEELQKQEIMKKFMSQHPEMDFSNAKLS 945
>CA860239 weakly similar to SP|P17624|NUDC_ Nuclear movement protein NUDC.
[Aspergillus nidulans] {Emericella nidulans}, partial
(24%)
Length = 271
Score = 71.2 bits (173), Expect = 4e-13
Identities = 40/87 (45%), Positives = 51/87 (57%)
Frame = +1
Query: 97 AGIKVNDEKSGVGTEKKDGESGLAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGF 156
A IK D S E+K+ + L + L Y W QTLQE+++ +PVP GT+
Sbjct: 13 ADIKKTDYDSMTPEEQKEHDRILREKEEAEQAALP-YKWKQTLQEVDITIPVPKGTRGRN 189
Query: 157 VICEIKKNHLKVGLKGQPPIIDGELYK 183
+I IKK L VGLKGQPPII+GEL K
Sbjct: 190 LIVTIKKKSLLVGLKGQPPIIEGELCK 270
>TC80668 similar to GP|18389274|gb|AAL67080.1 unknown protein {Arabidopsis
thaliana}, complete
Length = 691
Score = 71.2 bits (173), Expect = 4e-13
Identities = 43/159 (27%), Positives = 73/159 (45%), Gaps = 1/159 (0%)
Frame = +1
Query: 133 YSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDGELYKFIKADECYW 192
+ W QTL E+N+ + +P S C I+ H+++G+KG PP ++ +L +K D +W
Sbjct: 136 FEWDQTLDEVNIYINLPPNVHSKLFYCTIQSKHIELGIKGNPPFLNHDLTSPVKTDSSFW 315
Query: 193 SIEDQSTVSILLTKHDQMDWWKCLVKGDPLINTQKVEPESSKLGELDSETRMTVEKMMFD 252
++ED + I L K D+ W + G G+LD+
Sbjct: 316 TLED-DIMHITLNKRDKGQTWPSPILGQ---------------GQLDA------------ 411
Query: 253 QRQKSMGLPTSEELE-KQEMMKKFMSQHPNMDFSGAKLS 290
S +LE K+ M+++F ++P DFS A+ S
Sbjct: 412 ---------YSTDLEQKRLMLQRFQEENPGFDFSQAQFS 501
>AJ501735 similar to GP|18389274|gb unknown protein {Arabidopsis thaliana},
partial (95%)
Length = 599
Score = 37.0 bits (84), Expect = 0.009
Identities = 13/39 (33%), Positives = 23/39 (58%)
Frame = +1
Query: 133 YSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLK 171
+ W QTL E+N+ + +P S C I+ H+++G+K
Sbjct: 124 FEWDQTLDEVNIYINLPPNVHSKLFYCTIQSKHIELGIK 240
>TC77763 similar to GP|15215674|gb|AAK91382.1 AT4g27500/F27G19_100
{Arabidopsis thaliana}, partial (38%)
Length = 2297
Score = 33.9 bits (76), Expect = 0.075
Identities = 36/140 (25%), Positives = 57/140 (40%), Gaps = 4/140 (2%)
Frame = +1
Query: 14 PSSSSQPKPSKTIPFSS-TFDPSKPTAFLEKVFDFIA--KESTDFFDKDSAEKMVLSAVR 70
P + +P P +T+P T + A ++ ++F KE+ + + + R
Sbjct: 1432 PKEAPKPSPVETLPAQKETKSKGRDLAAIDDDYEFENPHKEAPAAKEPEIDPAKLKEMKR 1611
Query: 71 AAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKK-DGESGLAAPNQGNGMD 129
++ KAK A K K+A + AKAA + EK EKK +SG A Q
Sbjct: 1612 EEEIAKAKLAAERKKKMAEKAAAKAALKAQKEAEKKLKVLEKKAKKKSGAVATPQEEQEA 1791
Query: 130 LEKYSWTQTLQELNVNVPVP 149
E + Q + V P P
Sbjct: 1792 AEVEATEQEKVDDVVEAPAP 1851
>TC77010 similar to PIR|S59560|S59560 histone H1.41 - garden pea, partial
(82%)
Length = 1535
Score = 30.4 bits (67), Expect = 0.83
Identities = 37/143 (25%), Positives = 60/143 (41%), Gaps = 46/143 (32%)
Frame = +2
Query: 1 MAIISDYQDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIA--KESTD---- 54
+++ S+ Q T +P++++QPKP KT ++ P + E + + I KE T
Sbjct: 47 LSLSSESQVSTMSPNAAAQPKPKKT---AAAKKPLSHPTYAEMITEAIVSLKERTGSSQH 217
Query: 55 ----FFDKDSAE------KMVL----------------------------SAVRAA--KV 74
F ++ + K++L + V+AA
Sbjct: 218 AITKFIEEKHKDLSPTFRKLILLHLKKSVASGKLVKVKNSFKIAPAVAKTAPVKAAIAPA 397
Query: 75 KKAKAVAAEKAKIASQEKAKAAA 97
KKAKAV AK A++ KAKAAA
Sbjct: 398 KKAKAVTKPAAKAATKPKAKAAA 466
Score = 27.7 bits (60), Expect = 5.4
Identities = 30/97 (30%), Positives = 38/97 (38%), Gaps = 3/97 (3%)
Frame = +2
Query: 20 PKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVRAAKVK---K 76
P +KT P + P+K + K AK +T K A+ + AAK K K
Sbjct: 350 PAVAKTAPVKAAIAPAKKAKAVTKP---AAKAAT----KPKAKAAAVKPKAAAKPKAAAK 508
Query: 77 AKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKK 113
KAV KAK K AA V + V T K
Sbjct: 509 PKAVVKPKAKSVKATPVKKAAKKVVAKKPKSVKTPVK 619
>BQ147280 similar to PIR|C86333|C863 hypothetical protein AAF79914.1
[imported] - Arabidopsis thaliana, partial (10%)
Length = 666
Score = 30.0 bits (66), Expect = 1.1
Identities = 27/103 (26%), Positives = 40/103 (38%), Gaps = 16/103 (15%)
Frame = +2
Query: 8 QDETQTPSSSSQPKPSKTIPFS-----------STFDPSKPTAFLEKVFDFIAKESTDFF 56
QD+ SS +PK S T P S T S+ T+ K+ D
Sbjct: 113 QDDNDKISSLKKPKNSNTKPLSKGSKLKKLLKEETEPISRATSASRSKVKKELKDDDDDS 292
Query: 57 DKDSAEKMVLSAVRAAK-----VKKAKAVAAEKAKIASQEKAK 94
D+D +K + + K VKK K V E+ + ++ K K
Sbjct: 293 DEDDDDKPIAKKISKTKVVKEEVKKKKKVKKEEEVVVTETKVK 421
>BQ752218 similar to GP|19170914|emb hypothetical protein {Encephalitozoon
cuniculi}, partial (5%)
Length = 637
Score = 28.9 bits (63), Expect = 2.4
Identities = 24/61 (39%), Positives = 29/61 (47%), Gaps = 2/61 (3%)
Frame = -2
Query: 58 KDSAEKMVLSAVRAA--KVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDG 115
K AEK A A K KKA+ A +KAK +E K A K EK + EKK+
Sbjct: 600 KKEAEKKKKKAREEAAKKAKKAREAAYKKAKEEKKEAEKKAKEEKRQAEKK-IKEEKKEA 424
Query: 116 E 116
E
Sbjct: 423 E 421
Score = 26.9 bits (58), Expect = 9.2
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Frame = -2
Query: 57 DKDSAEKMVLSAVRAA--KVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKD 114
+K AEK R A K+K+ K A +KAK +E+ + A ++ + + E KD
Sbjct: 504 EKKEAEKKAKEEKRQAEKKIKEEKKEAEKKAKKDKKEQDRKDAESALSRQDTEDSKESKD 325
Query: 115 G 115
G
Sbjct: 324 G 322
>TC87973 weakly similar to PIR|D86282|D86282 protein F10B6.26 [imported] -
Arabidopsis thaliana, partial (82%)
Length = 1073
Score = 28.9 bits (63), Expect = 2.4
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Frame = +1
Query: 109 GTEKKDGESGLAAPNQGNGMDLEKYSW--TQTLQELNVNVPVPNGTKSGFVICEIKKNHL 166
GT++K+ E L + +G+GM K W +T+++ + + G V + K +
Sbjct: 331 GTQEKEIEVLLISAQKGSGMQFPKGGWEKDETMEQAALRETIEEAGVIGSVESNLGKWYY 510
Query: 167 KVGLKGQPPIIDGELYKFIKADE 189
K K QP + +G ++ + + E
Sbjct: 511 K--SKRQPTMHEGYMFPLLVSKE 573
>TC85720 similar to SP|P08283|H1_PEA Histone H1. [Garden pea] {Pisum
sativum}, partial (73%)
Length = 968
Score = 28.5 bits (62), Expect = 3.2
Identities = 23/65 (35%), Positives = 28/65 (42%)
Frame = +3
Query: 58 KDSAEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGES 117
K SAEK +A K K V A KAK + KAKA KV + V + K +
Sbjct: 336 KPSAEKKPAAA--KPKTKAVAKVTAVKAKPGPKPKAKAVVKTKVAAKAKPVAAKPKPAAA 509
Query: 118 GLAAP 122
AP
Sbjct: 510 KPKAP 524
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 28.5 bits (62), Expect = 3.2
Identities = 28/113 (24%), Positives = 40/113 (34%), Gaps = 19/113 (16%)
Frame = +1
Query: 12 QTPSSSSQPKPSKTIPFSSTFDP----------SKPTAFLEKVFDFIAKESTDFFDK--- 58
Q PS PS +PFSSTF P S P F + A +S ++
Sbjct: 250 QPPSLPLSSSPSFHLPFSSTFSPTDFLISPFFLSSPNVFASPTTEAFANQSFNWNKNSLG 429
Query: 59 ------DSAEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEK 105
D ++ LS K ++V + QE+ K K N+ K
Sbjct: 430 EEEQQGDKKDEKNLSDFSFPTQTKPESVFQSSTNMFQQEQTKKQDIWKFNEPK 588
>TC87710 similar to PIR|T08953|T08953 Cdk-activating protein kinase (EC
2.7.1.-) 1 [validated] - Arabidopsis thaliana, partial
(64%)
Length = 1738
Score = 28.1 bits (61), Expect = 4.1
Identities = 15/45 (33%), Positives = 23/45 (50%), Gaps = 6/45 (13%)
Frame = -2
Query: 107 GVGTEKKDGESGLAAPNQGNGMDLEKYSW------TQTLQELNVN 145
GVG GE G+ + G+GMD + W +Q L+++N N
Sbjct: 207 GVGAGADVGEDGVFGCDFGSGMDAPTFCWRIHVQFSQILKKINYN 73
>TC87997 similar to GP|12597886|gb|AAG60194.1 putative
phosphatidylinositol-4-phosphate 5-kinase {Oryza
sativa}, partial (38%)
Length = 1295
Score = 27.7 bits (60), Expect = 5.4
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +1
Query: 10 ETQTPSSSSQPKPSKTIPFSSTFDPSK 36
+T T + + Q KPS TIPF+S F K
Sbjct: 358 QTPTHNKTHQNKPSFTIPFTSKFQSFK 438
>AJ497431 similar to GP|9759308|dbj contains similarity to
transfactor~gene_id:MPH15.16 {Arabidopsis thaliana},
partial (31%)
Length = 638
Score = 27.3 bits (59), Expect = 7.0
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = -1
Query: 189 ECYWSIEDQSTVSILLTKH 207
+C WSIE+ +T +LL KH
Sbjct: 431 DCIWSIEEVTTCLLLLIKH 375
>TC87288 similar to PIR|S64423|S64423 probable membrane protein YGR115c -
yeast (Saccharomyces cerevisiae), partial (9%)
Length = 935
Score = 27.3 bits (59), Expect = 7.0
Identities = 24/94 (25%), Positives = 37/94 (38%)
Frame = +1
Query: 12 QTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVRA 71
Q P S SQP P+K + S S+ L K + D + D + VR
Sbjct: 205 QLPDSVSQPAPAKLV---SAMKGSR-KKHLGSQMKLSVKWAPDVY--DPVPTLSSHTVRT 366
Query: 72 AKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEK 105
K K++ +EK + +K + G D+K
Sbjct: 367 KKQHKSRIKKSEKKGVKKSQKGSYSKGCSSKDKK 468
>AW585396
Length = 555
Score = 27.3 bits (59), Expect = 7.0
Identities = 16/57 (28%), Positives = 32/57 (56%)
Frame = +1
Query: 218 KGDPLINTQKVEPESSKLGELDSETRMTVEKMMFDQRQKSMGLPTSEELEKQEMMKK 274
KG+ +IN + E ++SK+ ++ +T ++ FDQ K + + + ++E MKK
Sbjct: 343 KGEKIINELRAELKASKVEAETAKKAVTEQQQHFDQSTK---IKLEKIVAEKEEMKK 504
>AW691228 similar to PIR|H96592|H96 probable multispanning membrane protein
[imported] - Arabidopsis thaliana, partial (15%)
Length = 403
Score = 27.3 bits (59), Expect = 7.0
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 1/29 (3%)
Frame = +1
Query: 14 PSSSSQPKPSKTIPFSSTFD-PSKPTAFL 41
PS S QP P T SS+F PS P+ FL
Sbjct: 43 PSLSVQPPPPSTSSLSSSFSPPSIPSTFL 129
>TC84703 weakly similar to GP|20268746|gb|AAM14076.1 unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 770
Score = 26.9 bits (58), Expect = 9.2
Identities = 24/83 (28%), Positives = 34/83 (40%)
Frame = +2
Query: 41 LEKVFDFIAKESTDFFDKDSAEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIK 100
L++V A E+ F K V ++ K +AK EKA + KA+AAA
Sbjct: 404 LKRVEKLAAPETDSVF---RGFKAVATSSDRLKASRAKKALEEKAIEKEKRKAEAAA--- 565
Query: 101 VNDEKSGVGTEKKDGESGLAAPN 123
SG+ D + L PN
Sbjct: 566 -----SGIDWRSDDSDDELQKPN 619
>TC78819 similar to GP|9558421|dbj|BAB03357.1 ESTs AU097577(C62003)
C28669(C62003) correspond to a region of the predicted
gene.~Similar to, complete
Length = 1064
Score = 26.9 bits (58), Expect = 9.2
Identities = 11/41 (26%), Positives = 22/41 (52%)
Frame = +2
Query: 216 LVKGDPLINTQKVEPESSKLGELDSETRMTVEKMMFDQRQK 256
L + DP++N + E S+ +D + +EK++ D +K
Sbjct: 713 LCQSDPVLNDGQSRDEESRQAVIDGVRKRFIEKVVADHEEK 835
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.127 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,436,327
Number of Sequences: 36976
Number of extensions: 95340
Number of successful extensions: 551
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 532
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 543
length of query: 290
length of database: 9,014,727
effective HSP length: 95
effective length of query: 195
effective length of database: 5,502,007
effective search space: 1072891365
effective search space used: 1072891365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC134822.4


