

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
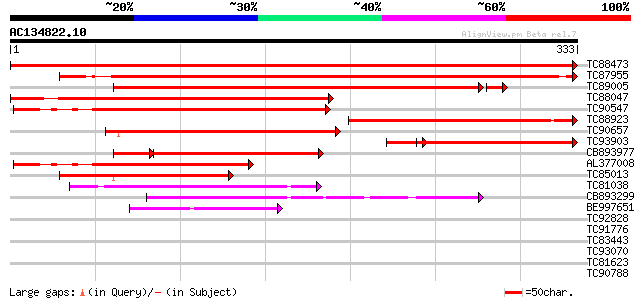
Score E
Sequences producing significant alignments: (bits) Value
TC88473 similar to PIR|T50560|T50560 SINAH1 protein [imported] -... 718 0.0
TC87955 similar to PIR|T47971|T47971 seven in absentia-like prot... 511 e-145
TC89005 similar to GP|22775634|dbj|BAC15488. similar to RING zin... 386 e-108
TC88047 similar to PIR|T50561|T50561 SINA1 protein [imported] - ... 345 1e-95
TC90547 similar to PIR|T50561|T50561 SINA1 protein [imported] - ... 271 2e-73
TC88923 homologue to GP|8468001|dbj|BAA96602.1 ESTs AU082689(E31... 268 2e-72
TC90657 similar to GP|22775634|dbj|BAC15488. similar to RING zin... 227 4e-60
TC93903 similar to GP|8468001|dbj|BAA96602.1 ESTs AU082689(E3198... 176 4e-53
CB893977 similar to GP|20136190|gb ring finger E3 ligase SINAT5 ... 176 4e-49
AL377008 homologue to GP|21536945|gb| seven in absentia-like pro... 187 6e-48
TC85013 similar to PIR|T50562|T50562 SINA2 protein [imported] - ... 124 4e-29
TC81038 weakly similar to GP|15864565|emb|CAC80703. SIAH1 protei... 95 3e-20
CB893299 weakly similar to GP|870794|gb|AA polyubiquitin {Arabid... 77 7e-15
BE997651 weakly similar to PIR|B96692|B966 hypothetical protein ... 62 4e-10
TC92828 weakly similar to GP|4115538|dbj|BAA36412.1 UDP-glycose:... 33 0.15
TC91776 similar to GP|18844905|dbj|BAB85374. putative zinc finge... 33 0.15
TC83443 similar to PIR|A86323|A86323 protein F14D16.3 [imported]... 32 0.26
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 32 0.26
TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcri... 32 0.34
TC90788 similar to PIR|F86340|F86340 protein F2D10.34 [imported]... 32 0.45
>TC88473 similar to PIR|T50560|T50560 SINAH1 protein [imported] - upland
cotton, partial (86%)
Length = 1360
Score = 718 bits (1854), Expect = 0.0
Identities = 332/333 (99%), Positives = 333/333 (99%)
Frame = +3
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA
Sbjct: 213 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 392
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK
Sbjct: 393 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 572
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR
Sbjct: 573 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 752
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMHTGCTFNHRYVKSNPREV+NATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 753 DDHKVDMHTGCTFNHRYVKSNPREVKNATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 932
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 933 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 1112
Query: 301 GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS
Sbjct: 1113GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 1211
>TC87955 similar to PIR|T47971|T47971 seven in absentia-like protein -
Arabidopsis thaliana, partial (83%)
Length = 1385
Score = 511 bits (1315), Expect = e-145
Identities = 239/305 (78%), Positives = 262/305 (85%), Gaps = 1/305 (0%)
Frame = +1
Query: 30 QQHHHHSEFSSLKPRSGGNNNHGVIGSTAIAPAT-SVHELLECPVCTNSMYPPIHQCHNG 88
+ HHH +FSS+ NN AP T SVH+LLECPVCTNSMYPPIHQCHNG
Sbjct: 415 EAEHHHHQFSSISKLL---NN---------APTTTSVHDLLECPVCTNSMYPPIHQCHNG 558
Query: 89 HTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKH 148
HTLCS CKTRVHNRCPTCRQELGDIRCLALEK+AESLE PC+Y SLGC EIFPY+SKLKH
Sbjct: 559 HTLCSNCKTRVHNRCPTCRQELGDIRCLALEKIAESLEFPCRYISLGCSEIFPYFSKLKH 738
Query: 149 ETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENA 208
E+IC FRPY CPYAGS+CS VG+I +LVAHLRDDH VDMH+GCTFNHRYVKSNP EVENA
Sbjct: 739 ESICTFRPYNCPYAGSDCSVVGNIPYLVAHLRDDHGVDMHSGCTFNHRYVKSNPTEVENA 918
Query: 209 TWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIW 268
TWMLTVFHCFGQYFCLHFEAFQL +PVYMAFLRFMGD+ +A+NY+YSLEVG NGRKL +
Sbjct: 919 TWMLTVFHCFGQYFCLHFEAFQLETSPVYMAFLRFMGDDRDAKNYSYSLEVGGNGRKLTF 1098
Query: 269 EGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCI 328
EG+PRSIRDSH+KV+DSHDGLII RNMALF SGGDRKELKLRVTGRIWKEQQN + C
Sbjct: 1099EGSPRSIRDSHKKVKDSHDGLIIYRNMALFXSGGDRKELKLRVTGRIWKEQQNSE---CT 1269
Query: 329 PNLCS 333
PN+CS
Sbjct: 1270PNMCS 1284
>TC89005 similar to GP|22775634|dbj|BAC15488. similar to RING zinc finger
protein {Oryza sativa (japonica cultivar-group)},
partial (73%)
Length = 926
Score = 386 bits (991), Expect(2) = e-108
Identities = 171/217 (78%), Positives = 192/217 (87%)
Frame = +1
Query: 62 ATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKV 121
++SV ELLECPVC N+MYPPIHQC NGHTLCS CK RVH+RCPTCR ELG+IRCLALEKV
Sbjct: 235 SSSVRELLECPVCLNAMYPPIHQCSNGHTLCSGCKPRVHDRCPTCRHELGNIRCLALEKV 414
Query: 122 AESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRD 181
A SLELPCKY GC I+PYYSKLKHE+ C FRPY CPYAGSEC+ VGD+ FLV HL+D
Sbjct: 415 AASLELPCKYQGFGCIGIYPYYSKLKHESQCVFRPYNCPYAGSECAVVGDVQFLVDHLKD 594
Query: 182 DHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFL 241
DHKVDMH+GCTFNHRYVKSNP+EVENATWMLTVF CFGQYFCLHFEAFQLGMAPVY+AFL
Sbjct: 595 DHKVDMHSGCTFNHRYVKSNPQEVENATWMLTVFSCFGQYFCLHFEAFQLGMAPVYIAFL 774
Query: 242 RFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDS 278
R MGD+++A+NYTYSLE G NGRK+ W+ PRSIR++
Sbjct: 775 RVMGDDDQAKNYTYSLEGGGNGRKMTWQEVPRSIREA 885
Score = 23.1 bits (48), Expect(2) = e-108
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +2
Query: 281 KVRDSHDGLIIQ 292
+VRDS DGLIIQ
Sbjct: 890 QVRDSFDGLIIQ 925
>TC88047 similar to PIR|T50561|T50561 SINA1 protein [imported] - Vitis
vinifera, partial (50%)
Length = 831
Score = 345 bits (886), Expect = 1e-95
Identities = 161/190 (84%), Positives = 168/190 (87%)
Frame = +3
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
M+LDSIECVSSSDGMDEDEI H HHHHSEFSS K R+GG N + ++G TAIA
Sbjct: 285 MELDSIECVSSSDGMDEDEI--------HSHHHHHSEFSSTKARNGGANINNILGPTAIA 440
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK
Sbjct: 441 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 620
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCKYYSLGCPEIFPYYSKLKHET CNFRPY+CPYAG S VGDI+FLVAHLR
Sbjct: 621 VAESLELPCKYYSLGCPEIFPYYSKLKHETECNFRPYSCPYAGIRVSTVGDISFLVAHLR 800
Query: 181 DDHKVDMHTG 190
DDH VDMHTG
Sbjct: 801 DDH*VDMHTG 830
>TC90547 similar to PIR|T50561|T50561 SINA1 protein [imported] - Vitis
vinifera, partial (42%)
Length = 572
Score = 271 bits (694), Expect = 2e-73
Identities = 132/186 (70%), Positives = 144/186 (76%)
Frame = +3
Query: 3 LDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIAPA 62
+DS VSS MDED LHPH +FSS + + S +
Sbjct: 81 MDSDSTVSSLIMMDED------LHPH--------QFSS--------STTSKLHSNGTPTS 194
Query: 63 TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVA 122
TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK+A
Sbjct: 195 TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKIA 374
Query: 123 ESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDD 182
ESLELPC+Y S+GCPEIFPYYSKLKHE+ICNFRPY CPYAGS+CS VGDI+ LVAHLRDD
Sbjct: 375 ESLELPCRYTSVGCPEIFPYYSKLKHESICNFRPYNCPYAGSDCSVVGDISQLVAHLRDD 554
Query: 183 HKVDMH 188
H+VDMH
Sbjct: 555 HRVDMH 572
>TC88923 homologue to GP|8468001|dbj|BAA96602.1 ESTs AU082689(E31988)
C99378(E10791) correspond to a region of the predicted
gene.~Similar to, partial (34%)
Length = 645
Score = 268 bits (686), Expect = 2e-72
Identities = 127/134 (94%), Positives = 130/134 (96%)
Frame = +2
Query: 200 SNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEV 259
SNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDEN+ARNY+YSLEV
Sbjct: 2 SNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENDARNYSYSLEV 181
Query: 260 GANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
GANGRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKE
Sbjct: 182 GANGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKE- 358
Query: 320 QNPDGGVCIPNLCS 333
QN D VCIPNLCS
Sbjct: 359 QNQDAAVCIPNLCS 400
>TC90657 similar to GP|22775634|dbj|BAC15488. similar to RING zinc finger
protein {Oryza sativa (japonica cultivar-group)},
partial (39%)
Length = 757
Score = 227 bits (579), Expect = 4e-60
Identities = 100/142 (70%), Positives = 115/142 (80%), Gaps = 4/142 (2%)
Frame = +3
Query: 57 TAIAPA----TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGD 112
TA+ P +SV ELLECPVC N+MYPPIHQC NGHT+CS CK RVHNRCPTCR ELG+
Sbjct: 330 TALKPTGTVLSSVRELLECPVCLNAMYPPIHQCSNGHTICSDCKPRVHNRCPTCRHELGN 509
Query: 113 IRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDI 172
IRCLALEKVA S LPCK+ GC I+PYY+K +HE+ C++RPY CPYAGSECS VGDI
Sbjct: 510 IRCLALEKVAASFALPCKFKDFGCIGIYPYYNKPEHESQCSYRPYNCPYAGSECSVVGDI 689
Query: 173 NFLVAHLRDDHKVDMHTGCTFN 194
N+LV HL++DHKVDMH G TFN
Sbjct: 690 NYLVTHLKEDHKVDMHNGSTFN 755
>TC93903 similar to GP|8468001|dbj|BAA96602.1 ESTs AU082689(E31988)
C99378(E10791) correspond to a region of the predicted
gene.~Similar to, partial (29%)
Length = 503
Score = 176 bits (446), Expect(2) = 4e-53
Identities = 82/94 (87%), Positives = 90/94 (95%)
Frame = +2
Query: 240 FLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFF 299
F RFMGDE +AR+Y+YSLEVG +GRKLI+EG+PRSIRDSH+KVRDSHDGLII RNMALFF
Sbjct: 56 FSRFMGDERDARSYSYSLEVGGSGRKLIYEGSPRSIRDSHKKVRDSHDGLIIYRNMALFF 235
Query: 300 SGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
SGGDRKELKLRVTGRIWKEQQNP+GGVCIPNLCS
Sbjct: 236 SGGDRKELKLRVTGRIWKEQQNPEGGVCIPNLCS 337
Score = 49.7 bits (117), Expect(2) = 4e-53
Identities = 22/24 (91%), Positives = 22/24 (91%)
Frame = +1
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMG 245
FCLHFEAFQLGMAPVYMAFL F G
Sbjct: 1 FCLHFEAFQLGMAPVYMAFLPFHG 72
>CB893977 similar to GP|20136190|gb ring finger E3 ligase SINAT5 {Arabidopsis
thaliana}, partial (37%)
Length = 353
Score = 176 bits (446), Expect(2) = 4e-49
Identities = 83/100 (83%), Positives = 86/100 (86%)
Frame = +1
Query: 85 CHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYS 144
C NG+TLCSTCKTRV NRC CRQELGDIRCLALEKVAESLEL CKYY LGC EIF YYS
Sbjct: 52 CKNGNTLCSTCKTRVDNRCRNCRQELGDIRCLALEKVAESLELRCKYYCLGCQEIFLYYS 231
Query: 145 KLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHK 184
KLKHETICNFR YT YAG EC VGDINF+VA+LRDDHK
Sbjct: 232 KLKHETICNFRLYTLCYAGLECYGVGDINFVVANLRDDHK 351
Score = 36.6 bits (83), Expect(2) = 4e-49
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = +2
Query: 62 ATSVHELLECPVCTNSMYPPIHQC 85
ATSVHELLEC VCTNS+ + C
Sbjct: 2 ATSVHELLECLVCTNSIARTVTHC 73
>AL377008 homologue to GP|21536945|gb| seven in absentia-like protein
{Arabidopsis thaliana}, partial (25%)
Length = 440
Score = 187 bits (474), Expect = 6e-48
Identities = 94/141 (66%), Positives = 102/141 (71%)
Frame = +3
Query: 3 LDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIAPA 62
+DS VSS MDED LHPH +FSS + + S +
Sbjct: 84 MDSDSTVSSLIMMDED------LHPH--------QFSS--------STTSKLHSNGTPTS 197
Query: 63 TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVA 122
TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK+A
Sbjct: 198 TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKIA 377
Query: 123 ESLELPCKYYSLGCPEIFPYY 143
ESLELPC+Y S+GCPEIFPYY
Sbjct: 378 ESLELPCRYTSVGCPEIFPYY 440
>TC85013 similar to PIR|T50562|T50562 SINA2 protein [imported] - Vitis
vinifera, partial (25%)
Length = 666
Score = 124 bits (312), Expect = 4e-29
Identities = 59/110 (53%), Positives = 73/110 (65%), Gaps = 8/110 (7%)
Frame = +1
Query: 30 QQHHHHSEFSSLKPRSGGNNNHGVIGSTAI--------APATSVHELLECPVCTNSMYPP 81
+ H S++ + K +S N + ST++ + VH+LL CPVC N MYPP
Sbjct: 334 ESHLLTSDYETGKAKSEAKINSTLTKSTSVGLNGKHGTSSKNGVHDLLGCPVCKNLMYPP 513
Query: 82 IHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKY 131
I+QC NGHTLCS CK VHN CPTC +LG+IRCLALEKVAESLELPC+Y
Sbjct: 514 IYQCPNGHTLCSNCKIEVHNLCPTCHHDLGNIRCLALEKVAESLELPCRY 663
>TC81038 weakly similar to GP|15864565|emb|CAC80703. SIAH1 protein {Brassica
napus}, partial (21%)
Length = 1016
Score = 95.1 bits (235), Expect = 3e-20
Identities = 48/148 (32%), Positives = 74/148 (49%)
Frame = +2
Query: 36 SEFSSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTC 95
+E +S+ +G NN S ++ S ++L+C +C+ + PI+QC NGH CS C
Sbjct: 197 NERNSVGSNAGQQNNDL---SKKVSAIISDPDVLDCFICSEPLAVPIYQCENGHIACSKC 367
Query: 96 KTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFR 155
+ N+CP C +G RC A+EK+ ES+++ C GC ++F K HE C +
Sbjct: 368 CGELRNKCPMCSMPIGYNRCRAVEKLLESIKISCPNAKYGCKDMFSCSMKSSHEKECIYI 547
Query: 156 PYTCPYAGSECSAVGDINFLVAHLRDDH 183
P CP+ G C + L H H
Sbjct: 548 PCKCPHTG--CGFLASSKELALHFSHRH 625
>CB893299 weakly similar to GP|870794|gb|AA polyubiquitin {Arabidopsis
thaliana}, partial (7%)
Length = 814
Score = 77.4 bits (189), Expect = 7e-15
Identities = 51/198 (25%), Positives = 83/198 (41%)
Frame = +2
Query: 81 PIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIF 140
P+ QC N H +CSTC ++ N C C + C +E +++S+++PC GC E
Sbjct: 5 PVFQCDNDHIVCSTCFPQLMNNCHKCSMPISSKCCKVIENISQSIQMPCPNKKYGCRETI 184
Query: 141 PYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKS 200
K KHE C + P CP G C V + L H H D ++ H + S
Sbjct: 185 SQSGKRKHEEECIYVPCYCPVKG--CDFVASLEVLSNHFNHKHG-DSLIEFSYGHSFTVS 355
Query: 201 NPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVG 260
E A + G+ F L+ LG A + + + Y+Y +
Sbjct: 356 LNSNDEAA---VLQEENDGKLFTLNNSTMLLGNA----VNISCIDVNSSEAGYSYDILAR 514
Query: 261 ANGRKLIWEGTPRSIRDS 278
+ +L + +P++I+ S
Sbjct: 515 SKTSRLKFHSSPKNIQRS 568
>BE997651 weakly similar to PIR|B96692|B966 hypothetical protein T12I7.7
[imported] - Arabidopsis thaliana, partial (8%)
Length = 267
Score = 61.6 bits (148), Expect = 4e-10
Identities = 30/91 (32%), Positives = 45/91 (48%), Gaps = 1/91 (1%)
Frame = +1
Query: 71 CPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESL-ELPC 129
C C + P+++C NGH +CSTC ++ +C C + RC A+E + S+ E+ C
Sbjct: 1 CSNCFELLTIPLYKCDNGHIVCSTCCDKLEKKCSKC--YIISKRCKAIENLLHSMEEISC 174
Query: 130 KYYSLGCPEIFPYYSKLKHETICNFRPYTCP 160
GC E Y KH+ C + P CP
Sbjct: 175 PNEKHGCRETIIYCITRKHDKECIYEPCYCP 267
>TC92828 weakly similar to GP|4115538|dbj|BAA36412.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (20%)
Length = 649
Score = 33.1 bits (74), Expect = 0.15
Identities = 37/164 (22%), Positives = 63/164 (37%), Gaps = 11/164 (6%)
Frame = +3
Query: 19 EIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGS-----TAIAPATSVHELLECPV 73
E+ I+H HHHH FS+ + G +H I S + P+ H L V
Sbjct: 72 ELAKLIIH-----HHHHQHFSNTILLTTGLMDHPSIDSYINRISTSHPSIIFHRLPSITV 236
Query: 74 ---CTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCK 130
T S +T+ K R + + + D+ C + + A S+ +P
Sbjct: 237 NHTTTQSRAATAFHFIKSNTVNVQSKLRQITQTSVIKAFIIDLFCTSAMETASSMLIPVY 416
Query: 131 YY---SLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGD 171
Y+ ++ Y+ K+ ET +F+ G E +A G+
Sbjct: 417 YFFTSGAAVLSLYSYFPKIHTETTISFK----DMVGVEIAAPGN 536
>TC91776 similar to GP|18844905|dbj|BAB85374. putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (27%)
Length = 1032
Score = 33.1 bits (74), Expect = 0.15
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Frame = +3
Query: 86 HNGHTLCSTCKTRVHNRCPTCRQELGDIRCL--ALEKVAESLELPCKY 131
H H+ C TR H CP C + LGD+ L+ + + ELP +Y
Sbjct: 624 HYMHSACFQAYTRSHYTCPICSKSLGDMAVYFGMLDALLAAEELPEEY 767
>TC83443 similar to PIR|A86323|A86323 protein F14D16.3 [imported] -
Arabidopsis thaliana, partial (21%)
Length = 1145
Score = 32.3 bits (72), Expect = 0.26
Identities = 17/52 (32%), Positives = 25/52 (47%), Gaps = 6/52 (11%)
Frame = +3
Query: 71 CPVCTNSMYP---PIHQCHNGHTLCSTC---KTRVHNRCPTCRQELGDIRCL 116
CP+C ++ P+ GH + STC T CP C + LGD++ L
Sbjct: 702 CPICHEYIFTSCSPVKALPCGHAMHSTCFKEYTCFSYTCPICSKSLGDMQVL 857
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 32.3 bits (72), Expect = 0.26
Identities = 14/35 (40%), Positives = 17/35 (48%)
Frame = -1
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHH 35
+ L SI C+ D + H LH HH HHHH
Sbjct: 394 LSLTSISCLWH*DQ*AQQIRHHHHLHHHHHHHHHH 290
>TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcription
factor [imported] - Arabidopsis thaliana, partial (47%)
Length = 847
Score = 32.0 bits (71), Expect = 0.34
Identities = 14/52 (26%), Positives = 24/52 (45%)
Frame = +1
Query: 8 CVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAI 59
C S M+ +QH H H Q HH + + R + H +IG++++
Sbjct: 118 CFFYSTSMEGHHLQHHQQHQHQQHQQHHQQH---QQRQHQQHAHNIIGTSSV 264
>TC90788 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana, partial (52%)
Length = 745
Score = 31.6 bits (70), Expect = 0.45
Identities = 17/52 (32%), Positives = 23/52 (43%), Gaps = 6/52 (11%)
Frame = +2
Query: 70 ECPVCTNSMYPP-----IHQCHNG-HTLCSTCKTRVHNRCPTCRQELGDIRC 115
EC +C + + QC +G H C H+ CP+CRQ L RC
Sbjct: 332 ECAICLSDFAAGDEIRVLPQCGHGFHVACIDTWLGSHSSCPSCRQILAVTRC 487
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,212,688
Number of Sequences: 36976
Number of extensions: 289688
Number of successful extensions: 3739
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 2895
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3385
length of query: 333
length of database: 9,014,727
effective HSP length: 97
effective length of query: 236
effective length of database: 5,428,055
effective search space: 1281020980
effective search space used: 1281020980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC134822.10


