

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.19 - phase: 0
(318 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
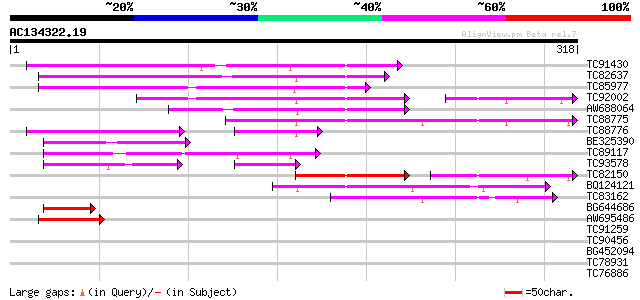
Score E
Sequences producing significant alignments: (bits) Value
TC91430 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 110 7e-25
TC82637 similar to PIR|G86331|G86331 IAA24 [imported] - Arabidop... 109 2e-24
TC85977 similar to GP|10176918|dbj|BAB10162. auxin response fact... 104 5e-23
TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response fact... 86 1e-22
AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsi... 97 8e-21
TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (... 91 6e-19
TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (... 60 6e-16
BE325390 weakly similar to GP|13272405|gb| unknown protein {Arab... 78 4e-15
TC89117 similar to GP|12484199|gb|AAG53998.1 auxin response tran... 77 1e-14
TC93578 similar to GP|9757747|dbj|BAB08228.1 auxin response fact... 55 4e-11
TC82150 similar to GP|19352051|dbj|BAB85919. auxin response fact... 64 6e-11
BQ124121 similar to PIR|G86331|G863 IAA24 [imported] - Arabidops... 51 5e-07
TC83162 homologue to PIR|F86427|F86427 auxin response factor 6 (... 50 1e-06
BG644686 similar to GP|22136676|gb| auxin response factor 1 {Ara... 45 3e-05
AW695486 homologue to GP|22136676|gb| auxin response factor 1 {A... 45 4e-05
TC91259 weakly similar to GP|17065054|gb|AAL32681.1 putative pro... 33 0.19
TC90456 weakly similar to PIR|G86466|G86466 hypothetical protein... 30 0.94
BG452094 weakly similar to PIR|T00443|T0 hypothetical protein T3... 29 2.1
TC78931 similar to PIR|E96802|E96802 unknown protein [imported] ... 28 3.6
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 28 4.7
>TC91430 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (23%)
Length = 728
Score = 110 bits (275), Expect = 7e-25
Identities = 69/217 (31%), Positives = 109/217 (49%), Gaps = 6/217 (2%)
Frame = +3
Query: 10 QQQQPSHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRP 69
Q+ + + ++W C G V +P SRV YFPQGH E + S++ + P
Sbjct: 99 QEGEKRVLDSELWHACAGPLVCLPADGSRVVYFPQGHSEQVAVSTNREVDTYIPNPSLPP 278
Query: 70 FTICIISAVDLLADPHTDEVFAKLLLTPVTNNSCVQD--PHEVPNCSNDDDVCDEVIDSF 127
IC + + + AD TDEV+A++ L P+ + P E+ + + + F
Sbjct: 279 QLICQLHNLTMHADTETDEVYAQMTLQPLNPEEQKEAYLPAELGTANK------QPTNYF 440
Query: 128 TRILALTNVSKH-AFYIPRFCAENMFPPLG---MEVSQHLLVTDVHGEVWKFHHVCHGFA 183
+IL ++ S H F +PR AE +FPPL +Q L+ D+HG WKF H+ G
Sbjct: 441 CKILTASDTSTHGGFSVPRRAAEKVFPPLDFTQQPPAQELIARDLHGNDWKFKHIFRGQP 620
Query: 184 KRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFV 220
KR++ T+ W+ FV K+L GD+V+F+ N F+
Sbjct: 621 KRHLL-TTGWSVFVSAKRLVAGDSVLFIWNEKISCFL 728
>TC82637 similar to PIR|G86331|G86331 IAA24 [imported] - Arabidopsis
thaliana, partial (28%)
Length = 889
Score = 109 bits (272), Expect = 2e-24
Identities = 63/202 (31%), Positives = 104/202 (51%), Gaps = 5/202 (2%)
Frame = +1
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPF-TICII 75
++ ++W C G V +P++ VYYFPQGH E ++S+ + P +C +
Sbjct: 130 LNSELWHACAGPLVSLPQVGGLVYYFPQGHSEQVAASTRRIATSQIPNYPSLPSQLLCQV 309
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTN 135
V L AD TDE++A++ L P+ + V E + + F + L ++
Sbjct: 310 QNVTLHADKETDEIYAQMTLQPLNSEKEVFPVSEFGLSKSKHPT-----EFFCKTLTASD 474
Query: 136 VSKH-AFYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTS 191
S H F +PR AE +FPPL + +Q L+V D+H W F H+ G KR++ T+
Sbjct: 475 TSTHGGFSVPRRAAEKLFPPLDYTIQPPTQELVVRDLHDNTWTFRHIYRGQPKRHLL-TT 651
Query: 192 EWASFVERKKLDVGDAVVFMKN 213
W+ FV K+L GD+V+F+++
Sbjct: 652 GWSLFVGSKRLRAGDSVLFIRD 717
>TC85977 similar to GP|10176918|dbj|BAB10162. auxin response factor-like
protein {Arabidopsis thaliana}, partial (65%)
Length = 3162
Score = 104 bits (259), Expect = 5e-23
Identities = 61/190 (32%), Positives = 98/190 (51%), Gaps = 4/190 (2%)
Frame = +3
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIIS 76
++ ++W C G V +P+ V+YFPQGH+E +S++ A + + RP +C +
Sbjct: 297 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRVI 476
Query: 77 AVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNV 136
V L A+P TDEVFA++ L P N QD + V + + SF + L ++
Sbjct: 477 NVMLKAEPDTDEVFAQVTLVPEPN----QDENAVEKEAPPAPPPRFHVHSFCKTLTASDT 644
Query: 137 SKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSE 192
S H F + + A+ PPL M +Q L+ D+HG W+F H+ G +R++ S
Sbjct: 645 STHGGFSVLKRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGQPRRHLL-QSG 821
Query: 193 WASFVERKKL 202
W+ FV K+L
Sbjct: 822 WSVFVSSKRL 851
>TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response factor 7
{Arabidopsis thaliana}, partial (31%)
Length = 1138
Score = 85.9 bits (211), Expect(2) = 1e-22
Identities = 51/157 (32%), Positives = 84/157 (53%), Gaps = 4/157 (2%)
Frame = +3
Query: 72 ICIISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRIL 131
IC++ V L ADP TDEV+A++ L PV D + + + F + L
Sbjct: 42 ICMLHNVALHADPETDEVYAQMTLQPVNK----YDKEAMLASDMGLKQNQQPSEFFCKTL 209
Query: 132 ALTNVSKH-AFYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHHVCHGFAKRNV 187
++ S H F +PR AE +FPPL + +Q ++ D++ W F H+ G KR++
Sbjct: 210 TASDTSTHGGFSVPRRAAEKIFPPLDFSMQPPAQEIVAKDLYDTTWTFRHIYRGQPKRHL 389
Query: 188 FYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRR 224
T+ W+ FV K+L GD+V+F+++ +L +GI+R
Sbjct: 390 L-TTGWSVFVSTKRLFAGDSVLFIRDEKQQLLLGIKR 497
Score = 37.7 bits (86), Expect(2) = 1e-22
Identities = 27/82 (32%), Positives = 40/82 (47%), Gaps = 8/82 (9%)
Frame = +1
Query: 245 LAEENKPFEIVYYPRGDDWCDFVVDGNIVDESM--KIQWNPRMRVKMKTDKSSRIPYQGT 302
LA N PF I Y PR C+FV+ ++++ + R R+ +T+ S Y GT
Sbjct: 583 LASNNSPFTIFYNPRTSP-CEFVIPLAKYNKALYTHVSLGMRFRMMFETEDSGVRRYMGT 759
Query: 303 ITTVSR------TSNLWRMLQV 318
IT +S ++ WR LQV
Sbjct: 760 ITGISDLDPVRWKNSQWRNLQV 825
>AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsis thaliana},
partial (15%)
Length = 576
Score = 97.1 bits (240), Expect = 8e-21
Identities = 52/139 (37%), Positives = 80/139 (57%), Gaps = 4/139 (2%)
Frame = +1
Query: 90 FAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNVSKHA-FYIPRFCA 148
F+K+ L P+ N+ D + N + SF + L ++ + F +PR+CA
Sbjct: 1 FSKITLIPLRNSELENDDSDGDGSENSEKPA-----SFAKTLTQSDANNGGGFSVPRYCA 165
Query: 149 ENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSEWASFVERKKLDVG 205
E +FP L Q ++ DVHGEVWKF H+ G +R++ T+ W+SFV +KKL G
Sbjct: 166 ETIFPRLDYSAEPPVQTVIAKDVHGEVWKFRHIYRGTPRRHLL-TTGWSSFVNQKKLVAG 342
Query: 206 DAVVFMKNSTGKLFVGIRR 224
D++VF++ +G+LFVGIRR
Sbjct: 343 DSIVFLRAESGELFVGIRR 399
>TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (23%)
Length = 678
Score = 90.9 bits (224), Expect = 6e-19
Identities = 66/218 (30%), Positives = 107/218 (48%), Gaps = 21/218 (9%)
Frame = +3
Query: 122 EVIDSFTRILALTNVSKHA-FYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHH 177
E + F + L ++ S H F +PR AE +FPPL +Q L+ D+HG WKF H
Sbjct: 9 EATNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRH 188
Query: 178 VCHGFAKRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ------- 230
+ G KR++ T+ W+ FV K+L GD+V+F+ N +L +GIRR +
Sbjct: 189 IFRGQPKRHLL-TTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVL 365
Query: 231 KKDELEKAVM-EAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESM---KIQWNPRMR 286
D + ++ A A N F I Y PR +FV+ +++ ++ R R
Sbjct: 366 SSDSMHLGLLAAAAHAAATNSRFTIFYNPRACP-SEFVIPLAKYVKAVYHTRVSVGMRFR 542
Query: 287 VKMKTDKSSRIPYQGTITTVSRTSNL------WRMLQV 318
+ +T++SS Y GTIT + +++ WR ++V
Sbjct: 543 MLFETEESSVRRYMGTITGICDLNSVRWPNSHWRSVKV 656
>TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (22%)
Length = 1278
Score = 60.1 bits (144), Expect(2) = 6e-16
Identities = 31/90 (34%), Positives = 48/90 (52%), Gaps = 1/90 (1%)
Frame = +3
Query: 10 QQQQPSHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEH-ASSSSSNAYIHSLDLQRFR 68
Q+ + + ++W C G V +P + SRV YFPQGH E A S++ H +
Sbjct: 552 QEGEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLP 731
Query: 69 PFTICIISAVDLLADPHTDEVFAKLLLTPV 98
P IC + + + AD TDEV+A++ L P+
Sbjct: 732 PQLICQLHNLTMHADVETDEVYAQMTLQPL 821
Score = 41.2 bits (95), Expect(2) = 6e-16
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 4/53 (7%)
Frame = +1
Query: 127 FTRILALTNVSKHA-FYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKF 175
F + L ++ S H F +PR AE +FPPL +Q L+ D+HG WKF
Sbjct: 895 FCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKF 1053
>BE325390 weakly similar to GP|13272405|gb| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 388
Score = 78.2 bits (191), Expect = 4e-15
Identities = 37/82 (45%), Positives = 49/82 (59%)
Frame = +3
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIISAVD 79
Q+W C GA VQ+P L+++V+YFPQGH EHA + R P C ISA+
Sbjct: 120 QLWHACAGAMVQMPPLNTKVFYFPQGHAEHAHNKV------DFSKTRVPPLIPCRISAMK 281
Query: 80 LLADPHTDEVFAKLLLTPVTNN 101
+ADP TDEV+ K+ LTP+ N
Sbjct: 282 YMADPETDEVYVKMKLTPLREN 347
>TC89117 similar to GP|12484199|gb|AAG53998.1 auxin response transcription
factor 3 {Arabidopsis thaliana}, partial (26%)
Length = 861
Score = 76.6 bits (187), Expect = 1e-14
Identities = 51/165 (30%), Positives = 78/165 (46%), Gaps = 10/165 (6%)
Frame = +1
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIISAVD 79
++W C G + +PK S V Y PQGH E A +A P C + V
Sbjct: 370 ELWHACAGPLISLPKKGSIVVYVPQGHFEQAHDFPVSAC-------NIPPHVFCRVLDVK 528
Query: 80 LLADPHTDEVFAKLLLTPVTNNSCVQDPHE-VPNCSNDDDVCDEVIDS-----FTRILAL 133
L A+ +DEV+ ++LL P N Q+ E V + +++ + ++ S F + L
Sbjct: 529 LHAEEGSDEVYCQVLLVP-ENQQLEQNVREGVIDADAEEEDTEAIVKSTTPHMFCKTLTA 705
Query: 134 TNVSKH-AFYIPRFCAENMFPPLG---MEVSQHLLVTDVHGEVWK 174
++ S H F +PR AE+ FPPL SQ L+ D+HG W+
Sbjct: 706 SDTSTHGGFSVPRXAAEDCFPPLDYGQQRPSQELVXKDLHGSKWE 840
>TC93578 similar to GP|9757747|dbj|BAB08228.1 auxin response factor 4
{Arabidopsis thaliana}, partial (18%)
Length = 634
Score = 55.5 bits (132), Expect(2) = 4e-11
Identities = 31/80 (38%), Positives = 42/80 (51%), Gaps = 2/80 (2%)
Frame = +1
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSS--SNAYIHSLDLQRFRPFTICIISA 77
++W C G +PK + V YFPQGHLE +S S I + DLQ P C +
Sbjct: 184 ELWHACAGPLTSLPKKGNVVVYFPQGHLEQFASFSPFKQLEIPNYDLQ---PQIFCRVVN 354
Query: 78 VDLLADPHTDEVFAKLLLTP 97
V LLA+ DEV+ ++ L P
Sbjct: 355 VQLLANKENDEVYTQVTLLP 414
Score = 29.3 bits (64), Expect(2) = 4e-11
Identities = 14/38 (36%), Positives = 23/38 (59%), Gaps = 1/38 (2%)
Frame = +2
Query: 127 FTRILALTNVSKHA-FYIPRFCAENMFPPLGMEVSQHL 163
F + L +++ S H F +PR AE+ FPPL ++ + L
Sbjct: 521 FCKTLTVSDTSTHGGFSVPRRAAEDCFPPLDYKLQRPL 634
>TC82150 similar to GP|19352051|dbj|BAB85919. auxin response factor 10
{Oryza sativa}, partial (30%)
Length = 1263
Score = 64.3 bits (155), Expect = 6e-11
Identities = 28/64 (43%), Positives = 43/64 (66%)
Frame = +1
Query: 161 QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFV 220
Q ++ D+HG+ WKF H+ G +R++ T+ W++FV KKL GD++VF++ G L V
Sbjct: 1 QTIIAKDMHGQCWKFKHIYRGTPRRHLL-TTGWSNFVNHKKLVAGDSIVFLRAENGDLCV 177
Query: 221 GIRR 224
GIRR
Sbjct: 178 GIRR 189
Score = 50.8 bits (120), Expect = 7e-07
Identities = 37/91 (40%), Positives = 52/91 (56%), Gaps = 9/91 (9%)
Frame = +2
Query: 237 KAVMEAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKM--KTDKS 294
++V+EAV A + FE VYYPR +F V + V +M+IQW MR KM +T+ S
Sbjct: 353 ESVVEAVNGAVNGRSFEGVYYPRAST-PEFCVKVSSVKSAMQIQWCSGMRFKMPFETEDS 529
Query: 295 SRIP-YQGTITTVSRTSNL------WRMLQV 318
SRI + GTI++V + WR+LQV
Sbjct: 530 SRISWFMGTISSVHVQDPIRWPDSPWRLLQV 622
>BQ124121 similar to PIR|G86331|G863 IAA24 [imported] - Arabidopsis thaliana,
partial (2%)
Length = 569
Score = 51.2 bits (121), Expect = 5e-07
Identities = 44/172 (25%), Positives = 75/172 (43%), Gaps = 16/172 (9%)
Frame = +2
Query: 148 AENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSEWASFVERKKLDV 204
A+ FP L M + Q L+V D+ G W F H+ + ++ T+ W +FV K L
Sbjct: 5 ADRCFP*LDMTLQPPMQDLVVKDLQGIEWNFRHIYCDHERAHLL-TNGWNTFVNSKNLRP 181
Query: 205 GDAVVFMKNSTGKLFVGIRR---------KDAAEQKKDELEKAVMEAVKLAEE-NKPFEI 254
GD+ +F+ G++ +GIRR +Q ++ + AV A F +
Sbjct: 182 GDSCIFVSGENGEIGIGIRRAMKQHSHICTKLCQQSSQNIQLGALAAVVHAVSIGSLFHL 361
Query: 255 VYYPRGDDWC---DFVVDGNIVDESMKIQWNPRMRVKMKTDKSSRIPYQGTI 303
Y+P W +F++ ES++ ++ RV M ++ GTI
Sbjct: 362 QYHP----WIAPFEFMIPLKTYVESIEKDYSIGTRVHMLSEVGGCPRRYGTI 505
>TC83162 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (18%)
Length = 569
Score = 49.7 bits (117), Expect = 1e-06
Identities = 38/142 (26%), Positives = 68/142 (47%), Gaps = 15/142 (10%)
Frame = +2
Query: 181 GFAKRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKD 233
G ++ + T+ W+ FV K+L GD+V+F+ N +L +GIRR + D
Sbjct: 41 GDSQNDTLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSD 220
Query: 234 ELEKAVM-EAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNP-------RM 285
+ ++ A A N F + + PR +FV+ + + +K ++ R
Sbjct: 221 SMHIGLLAAAAHAAATNSCFTVFFNPRASP-SEFVIP---LSKYIKAVYHTRPFLSVCRF 388
Query: 286 RVKMKTDKSSRIPYQGTITTVS 307
R+ +T++SS Y GTIT++S
Sbjct: 389 RMLFETEESSVRRYMGTITSIS 454
>BG644686 similar to GP|22136676|gb| auxin response factor 1 {Arabidopsis
thaliana}, partial (6%)
Length = 731
Score = 45.4 bits (106), Expect = 3e-05
Identities = 16/29 (55%), Positives = 21/29 (72%)
Frame = +3
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLE 48
++W+ C G V +PK RVYYFPQGH+E
Sbjct: 207 ELWRLCAGPLVDVPKNEERVYYFPQGHME 293
>AW695486 homologue to GP|22136676|gb| auxin response factor 1 {Arabidopsis
thaliana}, partial (9%)
Length = 480
Score = 45.1 bits (105), Expect = 4e-05
Identities = 16/37 (43%), Positives = 24/37 (64%)
Frame = +2
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSS 53
++ ++W C G V +P+ RVYYFPQGH+E +S
Sbjct: 299 LYKELWHACAGPLVTLPREGERVYYFPQGHMEQLEAS 409
>TC91259 weakly similar to GP|17065054|gb|AAL32681.1 putative protein
{Arabidopsis thaliana}, partial (6%)
Length = 723
Score = 32.7 bits (73), Expect = 0.19
Identities = 22/77 (28%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Frame = +2
Query: 4 NQYHHQQQQQPSHVHPQIW--QTCTGAAVQIP-KLHSRVYYFPQGHL-EHASSSSSNAYI 59
NQ+H+QQQQQ ++ +++ Q + +AV +P +H + F Q ++ + + S Y
Sbjct: 266 NQHHYQQQQQQQMLNSELYNVQMDSASAVPLPTTMHESMLPFYQSNVCDPNRADSGLTYN 445
Query: 60 HSLDLQRFRPFTICIIS 76
+ L +R R F+ ++S
Sbjct: 446 NPLQRKRSRDFSTELVS 496
>TC90456 weakly similar to PIR|G86466|G86466 hypothetical protein AAD39608.1
[imported] - Arabidopsis thaliana, partial (2%)
Length = 1332
Score = 30.4 bits (67), Expect = 0.94
Identities = 10/57 (17%), Positives = 31/57 (53%)
Frame = +3
Query: 250 KPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKMKTDKSSRIPYQGTITTV 306
+ F+ ++P+ + +C + + + + I+ PR+ + +KTD ++G ++++
Sbjct: 942 REFQKSFFPKFESFCSSITEHVPTADQLIIEEGPRLHIPLKTDSHIVSDFEGELSSI 1112
>BG452094 weakly similar to PIR|T00443|T0 hypothetical protein T30B22.29 -
Arabidopsis thaliana (fragment), partial (8%)
Length = 619
Score = 29.3 bits (64), Expect = 2.1
Identities = 13/37 (35%), Positives = 21/37 (56%)
Frame = +2
Query: 250 KPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMR 286
K F+++ P ++CDFV+ D +MK W+P R
Sbjct: 479 KSFKLIL-PELINFCDFVIXKTFYDAAMKRNWSPXXR 586
>TC78931 similar to PIR|E96802|E96802 unknown protein [imported] -
Arabidopsis thaliana, partial (85%)
Length = 661
Score = 28.5 bits (62), Expect = 3.6
Identities = 14/38 (36%), Positives = 21/38 (54%), Gaps = 7/38 (18%)
Frame = +2
Query: 157 MEVSQHLLVTDV-HGEVWKFHHV------CHGFAKRNV 187
+++ HL+ ++ HG VW+FH V CH RNV
Sbjct: 44 LKLELHLVNLELGHGRVWEFHDVFLSSIHCHPLTSRNV 157
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 28.1 bits (61), Expect = 4.7
Identities = 14/52 (26%), Positives = 24/52 (45%), Gaps = 6/52 (11%)
Frame = +2
Query: 4 NQYHHQQQQQPS------HVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEH 49
NQ HQQQQQ H + Q W + A + + ++ +PQ ++ +
Sbjct: 251 NQNQHQQQQQQQWAQQQPHQYQQQWMSMQYPATAMAMMQQQMMMYPQHYMPY 406
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,067,052
Number of Sequences: 36976
Number of extensions: 196251
Number of successful extensions: 1228
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1169
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1196
length of query: 318
length of database: 9,014,727
effective HSP length: 96
effective length of query: 222
effective length of database: 5,465,031
effective search space: 1213236882
effective search space used: 1213236882
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC134322.19


