

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
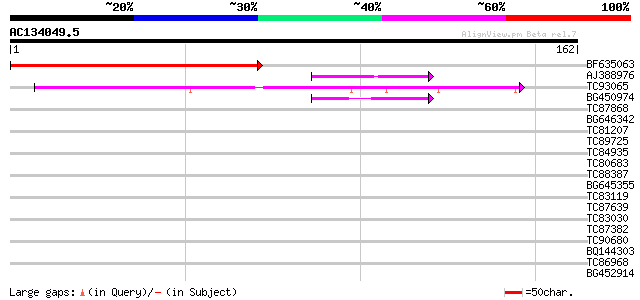
Query= AC134049.5 + phase: 0
(162 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 135 9e-33
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 40 4e-04
TC93065 40 5e-04
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 40 5e-04
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 39 0.001
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 39 0.001
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 39 0.001
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 38 0.002
TC84935 similar to PIR|G96631|G96631 probable RNA-binding protei... 37 0.003
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 34 0.029
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 32 0.11
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 31 0.19
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 31 0.19
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 31 0.25
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 30 0.32
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 30 0.55
TC90680 similar to GP|12836314|dbj|BAB23601. data source:SPTR s... 29 0.94
BQ144303 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 28 1.6
TC86968 similar to PIR|T07107|T07107 probable calcium-binding pr... 28 2.1
BG452914 similar to GP|13374061|emb ZF-HD homeobox protein {Flav... 27 2.7
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 135 bits (339), Expect = 9e-33
Identities = 68/72 (94%), Positives = 70/72 (96%)
Frame = -2
Query: 1 MEQLAEFNKILDDLENIEVQLEDEDKAILLLCALPRSFESFKDTMLYGKEGTVTLEEVQA 60
MEQL EFNKILDDLENIEVQLEDE+KAILLLCALP+SFESFKDTMLYGKEGTVTLEEVQA
Sbjct: 217 MEQLTEFNKILDDLENIEVQLEDEEKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQA 38
Query: 61 ALRTKELTKSKD 72
ALRTKELTKS D
Sbjct: 37 ALRTKELTKSND 2
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 40.0 bits (92), Expect = 4e-04
Identities = 18/35 (51%), Positives = 20/35 (56%)
Frame = +2
Query: 87 GNGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
G GGG G RG S G D KC+ C + GHF R C
Sbjct: 284 GGGGGGGGRGRSGGGGSD-LKCYXCGEPGHFARXC 385
>TC93065
Length = 783
Score = 39.7 bits (91), Expect = 5e-04
Identities = 39/163 (23%), Positives = 68/163 (40%), Gaps = 23/163 (14%)
Frame = +2
Query: 8 NKILDDLENIEVQLEDEDKAILLLCALPRSFESFKDTMLYGKE-GTVTLEEVQAALRTKE 66
+K++ + + L D+ +L LP FE+ ++ K +T+ E+ AL+ E
Sbjct: 284 SKVVTQIRLLGEDLSDQRVVEKILVCLPEMFEAKISSLEENKNFSEITVAELVNALQASE 463
Query: 67 LTKSKDLRADEDNEGLSVSKGNGGGRGNRG-----------SSKSGNKDKY-------KC 108
+S LR +E+ EG ++ G + + K + DK+ KC
Sbjct: 464 QRRS--LRMEENVEGAFLANNKGKNQSFKSFGKKKFPPCPHCKKDTHLDKFCWYRPGVKC 637
Query: 109 FKCHKLGHFKRDC---PEDNESSAQVVSEEYGDAGALV-VSCW 147
C++LGH ++ C E A+VV D L SC+
Sbjct: 638 RACNQLGHVEKVCKNKTNQQEQEARVVEHHQEDEEPLFKASCY 766
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 39.7 bits (91), Expect = 5e-04
Identities = 17/35 (48%), Positives = 21/35 (59%)
Frame = +1
Query: 87 GNGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
G GGGRG RG D KC++C + GHF R+C
Sbjct: 274 GRGGGRGGRGG------DDLKCYECGEPGHFAREC 360
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 38.9 bits (89), Expect = 0.001
Identities = 18/35 (51%), Positives = 22/35 (62%)
Frame = +3
Query: 87 GNGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
G GGGRG RG G D KC++C + GHF R+C
Sbjct: 279 GGGGGRG-RGGGGGGGSD-LKCYECGEPGHFAREC 377
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 38.9 bits (89), Expect = 0.001
Identities = 20/33 (60%), Positives = 24/33 (72%)
Frame = +2
Query: 2 EQLAEFNKILDDLENIEVQLEDEDKAILLLCAL 34
E L EFNKI+ DLENIEV LED A+++ C L
Sbjct: 185 ELLVEFNKIIGDLENIEVHLEDAG-ALMVWCCL 280
Score = 30.8 bits (68), Expect = 0.25
Identities = 15/25 (60%), Positives = 20/25 (80%), Gaps = 1/25 (4%)
Frame = +2
Query: 138 DAGALVV-SCWEEDEGEGSHLGIDS 161
DAGAL+V C E++EG+ SHLG D+
Sbjct: 248 DAGALMVWCCLEDEEGDVSHLGNDA 322
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 38.5 bits (88), Expect = 0.001
Identities = 20/59 (33%), Positives = 27/59 (44%)
Frame = +3
Query: 69 KSKDLRADEDNEGLSVSKGNGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDCPEDNES 127
K + L+ +DN G G GGGRG RG + C+ C GH RDC + +
Sbjct: 255 KGEPLQVRQDNHG-----GGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDCDRSDRN 410
Score = 31.2 bits (69), Expect = 0.19
Identities = 13/33 (39%), Positives = 16/33 (48%)
Frame = +3
Query: 89 GGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
GGG N G G C++C +GH RDC
Sbjct: 510 GGGNNNNGGGGYGGGGT-SCYRCGGVGHIARDC 605
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 38.1 bits (87), Expect = 0.002
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Frame = +1
Query: 87 GNGGGR-GNRGSSKSGNKDKYKCFKCHKLGHFKRDCP 122
G GGGR G G G C+ C + GHF RDCP
Sbjct: 70 GGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCP 180
>TC84935 similar to PIR|G96631|G96631 probable RNA-binding protein F8A5.17
[imported] - Arabidopsis thaliana, partial (41%)
Length = 552
Score = 37.4 bits (85), Expect = 0.003
Identities = 16/35 (45%), Positives = 22/35 (62%)
Frame = +2
Query: 88 NGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDCP 122
+ GGRG+ G+ +D CFKC + GH+ RDCP
Sbjct: 419 SSGGRGSYGAGDRVGQDD--CFKCGRPGHWARDCP 517
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 33.9 bits (76), Expect = 0.029
Identities = 25/75 (33%), Positives = 33/75 (43%), Gaps = 17/75 (22%)
Frame = +1
Query: 66 ELTKSKDLRADEDNEGLSVSKGNGGGRG---------------NRGS-SKSGNKDKYK-C 108
+L K K+ AD+D E S+ G ++G S SG D K C
Sbjct: 901 KLKKMKEKYADQDEEERSIRMSLLASSGKPIKKEETLPVIETSDKGKKSDSGPIDAPKIC 1080
Query: 109 FKCHKLGHFKRDCPE 123
+KC K+GH RDC E
Sbjct: 1081YKCKKVGHLSRDCKE 1125
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 32.0 bits (71), Expect = 0.11
Identities = 13/33 (39%), Positives = 15/33 (45%)
Frame = +1
Query: 89 GGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
GGG RG + G C C + GH RDC
Sbjct: 856 GGGGSLRGGYRDGGFRDVVCRSCQQFGHMSRDC 954
Score = 29.6 bits (65), Expect = 0.55
Identities = 10/17 (58%), Positives = 10/17 (58%)
Frame = +1
Query: 108 CFKCHKLGHFKRDCPED 124
C C K GH RDCP D
Sbjct: 730 CNNCRKTGHLARDCPND 780
Score = 26.6 bits (57), Expect = 4.7
Identities = 10/28 (35%), Positives = 14/28 (49%)
Frame = +1
Query: 95 RGSSKSGNKDKYKCFKCHKLGHFKRDCP 122
R S+ G C C + GH+ R+CP
Sbjct: 385 RRDSRRGFSQDNLCKNCKRPGHYVRECP 468
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 31.2 bits (69), Expect = 0.19
Identities = 20/58 (34%), Positives = 26/58 (44%)
Frame = -1
Query: 89 GGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDCPEDNESSAQVVSEEYGDAGALVVSC 146
GG N S G K C+KC + GH+ +CP + S+A VS G A C
Sbjct: 552 GGAYVNTVSGSGGASGK--CYKCQQPGHWASNCP--SMSAANRVSGGSGGASGNCYKC 391
Score = 30.8 bits (68), Expect = 0.25
Identities = 14/39 (35%), Positives = 20/39 (50%)
Frame = -1
Query: 84 VSKGNGGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDCP 122
VS G+GG GN C+KC++ GH+ +CP
Sbjct: 435 VSGGSGGASGN-------------CYKCNQPGHWANNCP 358
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 31.2 bits (69), Expect = 0.19
Identities = 20/78 (25%), Positives = 31/78 (39%), Gaps = 20/78 (25%)
Frame = +3
Query: 65 KELTKSKDLRADEDNEGLSVSKGNGGGRGN--------------------RGSSKSGNKD 104
KE+ K K+L+ D+ S S+ R R SS+ ++D
Sbjct: 162 KEVKKEKNLKMSSDSRSRSRSRSRSRSRSRSPRIRKIRSDRHSYRDAPYRRDSSRGFSRD 341
Query: 105 KYKCFKCHKLGHFKRDCP 122
C C + GH+ R+CP
Sbjct: 342 NL-CKNCKRPGHYARECP 392
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 30.8 bits (68), Expect = 0.25
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 4/68 (5%)
Frame = -1
Query: 67 LTKSKDLRADEDNEGLSVSKGNGGGRGNRGSSKSG----NKDKYKCFKCHKLGHFKRDCP 122
L SKD D G + G + SS G +D +C+ CH+ GH R+C
Sbjct: 451 LPVSKDRDGDMMMTGAIHKQRRRRGSSSSVSSAEGAPPPRRDMRECYSCHERGHIARNC- 275
Query: 123 EDNESSAQ 130
N S+A+
Sbjct: 274 -TNTSAAK 254
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 30.4 bits (67), Expect = 0.32
Identities = 13/34 (38%), Positives = 16/34 (46%)
Frame = +1
Query: 89 GGGRGNRGSSKSGNKDKYKCFKCHKLGHFKRDCP 122
GG + +R SS CF C + GH DCP
Sbjct: 109 GGRQSSRSSSSPNRSFAGTCFTCGESGHRASDCP 210
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 29.6 bits (65), Expect = 0.55
Identities = 16/44 (36%), Positives = 21/44 (47%)
Frame = +2
Query: 92 RGNRGSSKSGNKDKYKCFKCHKLGHFKRDCPEDNESSAQVVSEE 135
R R + K N + KCF C GH DC N+ +V+EE
Sbjct: 998 RPQRNTIKERNTNIPKCFICQGYGHIALDCV--NQKVFTIVNEE 1123
>TC90680 similar to GP|12836314|dbj|BAB23601. data source:SPTR source
key:Q9H0W3 evidence:ISS~homolog to HYPOTHETICAL 64.6
KDA PROTEIN~putative, partial (5%)
Length = 1280
Score = 28.9 bits (63), Expect = 0.94
Identities = 10/28 (35%), Positives = 15/28 (52%)
Frame = +2
Query: 94 NRGSSKSGNKDKYKCFKCHKLGHFKRDC 121
+R S + Y+C+KC + GH DC
Sbjct: 935 HRWSKTRSSLSTYECWKCQRPGHLAEDC 1018
>BQ144303 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (11%)
Length = 1158
Score = 28.1 bits (61), Expect = 1.6
Identities = 14/36 (38%), Positives = 18/36 (49%)
Frame = +1
Query: 78 DNEGLSVSKGNGGGRGNRGSSKSGNKDKYKCFKCHK 113
DN L++ G GG G G GN+ K KC+K
Sbjct: 109 DNIRLNMRPGGGGS*GGEGV*GGGNQAKQSVKKCYK 216
>TC86968 similar to PIR|T07107|T07107 probable calcium-binding protein
(clone Y8) - potato, partial (89%)
Length = 1691
Score = 27.7 bits (60), Expect = 2.1
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 8/70 (11%)
Frame = +1
Query: 80 EGLSVSKGNGGGRGNR-----GSSKS---GNKDKYKCFKCHKLGHFKRDCPEDNESSAQV 131
E SV + +GGG GN GS+ GNKD + F HK RD +D + S
Sbjct: 139 ESGSVKRKSGGGGGNAVEVLDGSNIMELVGNKDVFSNFVNHKFQELDRD--KDGKLSLTE 312
Query: 132 VSEEYGDAGA 141
+ D G+
Sbjct: 313 LQPAVSDIGS 342
>BG452914 similar to GP|13374061|emb ZF-HD homeobox protein {Flaveria
bidentis}, partial (29%)
Length = 454
Score = 27.3 bits (59), Expect = 2.7
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = +2
Query: 74 RADEDNEGLSVSKGNGGGRGNRGSSKSGNKDKYK 107
R D+D S S G GGG G GS SG++ +++
Sbjct: 209 REDDDMSNPSSSGGGGGGGG--GSGGSGSRKRFR 304
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.132 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,218,094
Number of Sequences: 36976
Number of extensions: 48904
Number of successful extensions: 333
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 317
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 330
length of query: 162
length of database: 9,014,727
effective HSP length: 89
effective length of query: 73
effective length of database: 5,723,863
effective search space: 417841999
effective search space used: 417841999
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC134049.5


