

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.18 - phase: 0
(166 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
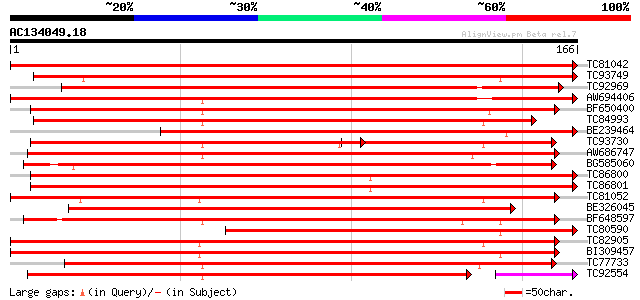
Score E
Sequences producing significant alignments: (bits) Value
TC81042 weakly similar to GP|16944811|emb|CAC82811. resistance g... 335 6e-93
TC93749 similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum tuber... 208 7e-55
TC92969 weakly similar to GP|20339360|gb|AAM19353.1 TIR-similar-... 190 3e-49
AW694406 weakly similar to GP|3947735|emb NL27 {Solanum tuberosu... 182 7e-47
BF650400 weakly similar to GP|9965109|gb| resistance protein MG1... 179 6e-46
TC84993 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 165 9e-42
BE239464 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 162 4e-41
TC93730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 112 9e-41
AW686747 weakly similar to PIR|A54810|A5 TMV resistance protein ... 161 1e-40
BG585060 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 156 3e-39
TC86800 weakly similar to GP|10944739|emb|CAC14089. hypothetical... 155 7e-39
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 155 7e-39
TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 150 2e-37
BE326045 similar to PIR|F96753|F96 Similar to downy mildew resis... 150 2e-37
BF648597 weakly similar to GP|12003378|gb Avr9/Cf-9 rapidly elic... 149 4e-37
TC80590 similar to GP|23477201|emb|CAD36199. NLS-TIR-NBS disease... 148 9e-37
TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 148 9e-37
BI309457 weakly similar to GP|15787901|gb resistance gene analog... 148 9e-37
TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 147 1e-36
TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance ... 139 3e-36
>TC81042 weakly similar to GP|16944811|emb|CAC82811. resistance gene-like
{Solanum tuberosum subsp. andigena}, partial (6%)
Length = 1016
Score = 335 bits (858), Expect = 6e-93
Identities = 165/166 (99%), Positives = 165/166 (99%)
Frame = +1
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGD 60
MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNN TSHLNGALKRLDIRTYIDNNLNSGD
Sbjct: 70 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNCTSHLNGALKRLDIRTYIDNNLNSGD 249
Query: 61 EISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDV 120
EISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDV
Sbjct: 250 EISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDV 429
Query: 121 RNQRGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 166
RNQRGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR
Sbjct: 430 RNQRGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 567
>TC93749 similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum tuberosum},
partial (17%)
Length = 772
Score = 208 bits (530), Expect = 7e-55
Identities = 102/161 (63%), Positives = 129/161 (79%), Gaps = 2/161 (1%)
Frame = +2
Query: 8 STSSSNNTPQEKH-EVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTL 66
S S+S +KH +VF+SFR EDTR FTSHL+ L RL + TYID NL GDEIS+TL
Sbjct: 11 SWSTSFPPHHQKHGQVFLSFRGEDTRYTFTSHLHATLTRLKVGTYIDYNLQRGDEISSTL 190
Query: 67 VRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGS 126
+ AIEEA++S+++FSKNY SK+CLDEL+KILECK+MKG++++PIFYD+DP+ VRNQ GS
Sbjct: 191 LMAIEEAKVSIVIFSKNYGNSKWCLDELVKILECKKMKGQILLPIFYDIDPSHVRNQTGS 370
Query: 127 YAEAFAKHEKNSEEKI-KVQEWRNGLMEAANYSGWDCNVNR 166
YAEAF KHEK + K+ KVQ WR+ L EAAN SGW+C+VNR
Sbjct: 371 YAEAFVKHEKQFQGKLEKVQTWRHALREAANISGWECSVNR 493
>TC92969 weakly similar to GP|20339360|gb|AAM19353.1
TIR-similar-domain-containing protein TSDC {Pisum
sativum}, partial (15%)
Length = 772
Score = 190 bits (482), Expect = 3e-49
Identities = 91/147 (61%), Positives = 118/147 (79%)
Frame = +3
Query: 16 PQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAEL 75
PQ+ H+VF+SFR EDTR FTSHL AL RL I+TYIDN L GDEIS +L++AI++A+L
Sbjct: 84 PQQIHDVFLSFRGEDTRYTFTSHLYAALTRLQIKTYIDNELERGDEISPSLLKAIDDAKL 263
Query: 76 SVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHE 135
SVI+FS+NYA+S++CL+EL+KILECK+ G+++VPIFY V+PT+VRNQ GSY + A+ E
Sbjct: 264 SVIIFSENYASSRWCLEELVKILECKKNNGQILVPIFYHVNPTNVRNQTGSYGLSLAEQE 443
Query: 136 KNSEEKIKVQEWRNGLMEAANYSGWDC 162
K + KVQ WR L EAAN+SGWDC
Sbjct: 444 KR-RDMHKVQTWRLALTEAANFSGWDC 521
>AW694406 weakly similar to GP|3947735|emb NL27 {Solanum tuberosum}, partial
(14%)
Length = 628
Score = 182 bits (461), Expect = 7e-47
Identities = 87/167 (52%), Positives = 126/167 (75%), Gaps = 1/167 (0%)
Frame = +3
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSG 59
M + +S+SS+ +VFISFR +DTR FTSHLN ALK+ ++T+ID++ L G
Sbjct: 54 MQINIGASSSSTLEVASNSFDVFISFRGDDTRRKFTSHLNEALKKSGVKTFIDDSELKKG 233
Query: 60 DEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTD 119
DEIS+ L++AIEE+ S+++FS++YA+SK+CL+EL+KILECK+ G++V+PIFY++DP+
Sbjct: 234 DEISSALIKAIEESCASIVIFSEDYASSKWCLNELVKILECKKDNGQIVIPIFYEIDPSH 413
Query: 120 VRNQRGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 166
VRNQ GSY +AFAKHEKN +K Q+W++ L E +N SGWD +R
Sbjct: 414 VRNQIGSYGQAFAKHEKN----LKQQKWKDALTEVSNLSGWDSKSSR 542
>BF650400 weakly similar to GP|9965109|gb| resistance protein MG13 {Glycine
max}, partial (36%)
Length = 644
Score = 179 bits (453), Expect = 6e-46
Identities = 85/157 (54%), Positives = 119/157 (75%), Gaps = 2/157 (1%)
Frame = +1
Query: 7 SSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTT 65
SS SSS+ PQ+K++VF+SFR +DTR NFTSHL L R + T+IDNN L GDEIS
Sbjct: 112 SSVSSSSVAPQKKYDVFLSFRGDDTRRNFTSHLYDTLSRKKVETFIDNNELEKGDEISPA 291
Query: 66 LVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRG 125
L++AIEE+ +S+I+FS+NYA+SK+CL+EL KIL+CK+ ++V+P+FY++DP+ VR Q G
Sbjct: 292 LIKAIEESHVSIIIFSENYASSKWCLNELKKILQCKKYMQQIVIPVFYNIDPSHVRKQTG 471
Query: 126 SYAEAFAKHEKNSE-EKIKVQEWRNGLMEAANYSGWD 161
SY +AF KH ++ + K+QEW+ L EAA+ GWD
Sbjct: 472 SYEQAFTKHMRDLKLNNDKLQEWKAALAEAASLVGWD 582
>TC84993 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (21%)
Length = 672
Score = 165 bits (417), Expect = 9e-42
Identities = 80/149 (53%), Positives = 115/149 (76%), Gaps = 2/149 (1%)
Frame = +2
Query: 8 STSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTL 66
S+SS+ +VFISFR +DTR FTSHLN ALK+ ++T+ID+N L GDEIS+ L
Sbjct: 80 SSSSTLEVASNSFDVFISFRGDDTRRKFTSHLNEALKKSGLKTFIDDNELKKGDEISSAL 259
Query: 67 VRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGS 126
++AIEE+ S+++ S+NYA+SK+CL+EL+KILECK+ G++V+PIFY++DP+ VR Q GS
Sbjct: 260 IKAIEESCASIVILSENYASSKWCLNELVKILECKKDNGQIVIPIFYEIDPSHVRYQIGS 439
Query: 127 YAEAFAKHEKN-SEEKIKVQEWRNGLMEA 154
Y +AFAK+EKN +K +Q+W++ L EA
Sbjct: 440 YGQAFAKYEKNLRHKKDNLQKWKDALTEA 526
>BE239464 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(9%)
Length = 561
Score = 162 bits (411), Expect = 4e-41
Identities = 74/123 (60%), Positives = 101/123 (81%), Gaps = 1/123 (0%)
Frame = +3
Query: 45 RLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMK 104
R DI+TYID+NL GDEIS L++AI+EA+LSVIVFSKNYA SK+CLDE++ IL C++ K
Sbjct: 3 RFDIKTYIDDNLERGDEISQALLKAIDEAKLSVIVFSKNYATSKWCLDEVVNILACRKNK 182
Query: 105 GKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNSEEKIKV-QEWRNGLMEAANYSGWDCN 163
G++++P+FY+VDP VR+Q GSYAEAF KHE+ + + Q+W++ L EAAN+SGWDC+
Sbjct: 183 GQIILPVFYEVDPFHVRHQLGSYAEAFVKHEQRFASTMNIAQKWKDALGEAANHSGWDCS 362
Query: 164 VNR 166
+NR
Sbjct: 363 INR 371
>TC93730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (68%)
Length = 690
Score = 112 bits (281), Expect(2) = 9e-41
Identities = 56/100 (56%), Positives = 76/100 (76%), Gaps = 2/100 (2%)
Frame = +2
Query: 7 SSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTT 65
SS SSS+ P +K++VF+SFR EDTR NFTSHL AL R + T+IDNN L GDEIS
Sbjct: 152 SSISSSDVVPLKKYDVFLSFRGEDTRRNFTSHLYDALSRKKVETFIDNNKLQKGDEISPA 331
Query: 66 LVRAIEEAELSVIVFSKNYAASKFCLDELM-KILECKRMK 104
L++AIE++ S+++FS+NYA+SK+CL+E + LECK+ K
Sbjct: 332 LIKAIEKSHASIVIFSENYASSKWCLNEAQERFLECKKYK 451
Score = 70.5 bits (171), Expect(2) = 9e-41
Identities = 30/64 (46%), Positives = 45/64 (69%), Gaps = 1/64 (1%)
Frame = +3
Query: 98 LECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN-SEEKIKVQEWRNGLMEAAN 156
L + ++ +V+P+FY +DP+ VRNQ GSY +AFAKH+++ K+QEW+ L EAA+
Sbjct: 432 LNARNIRNHIVIPVFYKIDPSHVRNQTGSYEQAFAKHKRDLKSNNDKLQEWKAALTEAAS 611
Query: 157 YSGW 160
SGW
Sbjct: 612 LSGW 623
>AW686747 weakly similar to PIR|A54810|A5 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (10%)
Length = 644
Score = 161 bits (407), Expect = 1e-40
Identities = 83/158 (52%), Positives = 112/158 (70%), Gaps = 2/158 (1%)
Frame = +3
Query: 6 SSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEIST 64
SSS+S S T + + VF+SFR EDTR FT HL +L+R I+T+ D++ L G+ IS
Sbjct: 78 SSSSSPSIQTSRWTNHVFLSFRGEDTRQGFTDHLFASLERRGIKTFKDDHDLERGEVISY 257
Query: 65 TLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQR 124
L +AIEE+ ++I+ S NYA+S +CLDEL KI+EC + G+ V PIFY VDP+DVR+QR
Sbjct: 258 ELNKAIEESMFAIIILSPNYASSTWCLDELKKIVECSKSFGQAVFPIFYGVDPSDVRHQR 437
Query: 125 GSYAEAFAKH-EKNSEEKIKVQEWRNGLMEAANYSGWD 161
GS+ EAF KH EK +++ KV+ WR+ L E A YSGWD
Sbjct: 438 GSFDEAFRKHEEKFRKDRTKVERWRDALREVAGYSGWD 551
>BG585060 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(9%)
Length = 817
Score = 156 bits (395), Expect = 3e-39
Identities = 78/157 (49%), Positives = 114/157 (71%), Gaps = 1/157 (0%)
Frame = +1
Query: 5 YSSSTSSSNNTPQ-EKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEIS 63
YSS S PQ +KH+VF+SFR D R F SHL AL R +I ++DN L GDEI+
Sbjct: 70 YSSEFQCS--VPQSDKHDVFVSFRGLDIREGFLSHLVEALSRKEIVFFVDNKLRKGDEIA 243
Query: 64 TTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQ 123
+L AIE + +S+++FS+NYA+SK+CLDEL+KI+EC+ G++++P+FY VDPT VR+Q
Sbjct: 244 QSLFEAIETSSISLVIFSQNYASSKWCLDELVKIVECREKDGQILLPVFYKVDPTVVRHQ 423
Query: 124 RGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGW 160
+G+YA AFA+HE+ KV++WR+ L ++A+ SG+
Sbjct: 424 KGTYANAFAEHEQKYNLN-KVKQWRSSLKKSADISGF 531
>TC86800 weakly similar to GP|10944739|emb|CAC14089. hypothetical protein
{Arabidopsis thaliana}, partial (8%)
Length = 672
Score = 155 bits (392), Expect = 7e-39
Identities = 75/163 (46%), Positives = 112/163 (68%), Gaps = 3/163 (1%)
Frame = +1
Query: 7 SSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTL 66
S SSS++ Q+K++VFISFR EDTR FTSHL+ AL R + TYID + GDE+ L
Sbjct: 31 SLASSSHSAAQKKYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYIDYRIEKGDEVWPEL 210
Query: 67 VRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMK---GKMVVPIFYDVDPTDVRNQ 123
+AI+++ L ++VFS+NYA+S +CL+EL++++EC+ V+P+FY VDP+ VR Q
Sbjct: 211 EKAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPVFYHVDPSHVRKQ 390
Query: 124 RGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 166
GSY A AKH++ +++ +Q W+N L +AAN SG+ + R
Sbjct: 391 TGSYGSALAKHKQENQDDKMMQNWKNALFQAANLSGFHSSTYR 519
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 155 bits (392), Expect = 7e-39
Identities = 75/163 (46%), Positives = 112/163 (68%), Gaps = 3/163 (1%)
Frame = +2
Query: 7 SSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTL 66
S SSS++ Q+K++VFISFR EDTR FTSHL+ AL R + TYID + GDE+ L
Sbjct: 29 SLASSSHSAAQKKYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYIDYRIEKGDEVWPEL 208
Query: 67 VRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMK---GKMVVPIFYDVDPTDVRNQ 123
+AI+++ L ++VFS+NYA+S +CL+EL++++EC+ V+P+FY VDP+ VR Q
Sbjct: 209 EKAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPVFYHVDPSHVRKQ 388
Query: 124 RGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 166
GSY A AKH++ +++ +Q W+N L +AAN SG+ + R
Sbjct: 389 TGSYGSALAKHKQENQDDKMMQNWKNALFQAANLSGFHSSTYR 517
>TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein
N {Arabidopsis thaliana}, partial (8%)
Length = 656
Score = 150 bits (380), Expect = 2e-37
Identities = 77/165 (46%), Positives = 112/165 (67%), Gaps = 4/165 (2%)
Frame = +2
Query: 1 MAWSYSSSTSSSNNTPQEK--HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDN-NLN 57
MA S +SS+S+ P+ K ++VF++FR EDTR NF HL AL+R I + D+ NL
Sbjct: 47 MASSSNSSSSALMALPRRKNYYDVFVTFRGEDTRFNFIDHLFAALQRKGIFAFRDDANLQ 226
Query: 58 SGDEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDP 117
G+ I L+RAIE +++ + V SKNY++S +CL EL+ IL+C ++ G+ V+P+FYDVDP
Sbjct: 227 KGESIPPELIRAIEGSQVFIAVLSKNYSSSTWCLRELVHILDCSQVSGRRVLPVFYDVDP 406
Query: 118 TDVRNQRGSYAEAFAKHEKN-SEEKIKVQEWRNGLMEAANYSGWD 161
++VR+Q+G Y EAF+KHE+ + VQ WR L + N SGWD
Sbjct: 407 SEVRHQKGIYGEAFSKHEQTFQHDSHVVQSWREALTQVGNISGWD 541
>BE326045 similar to PIR|F96753|F96 Similar to downy mildew resistance
protein RPP5 [imported] - Arabidopsis thaliana, partial
(6%)
Length = 587
Score = 150 bits (379), Expect = 2e-37
Identities = 72/131 (54%), Positives = 98/131 (73%)
Frame = +2
Query: 18 EKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSV 77
+K++VFISFR EDTR+NFTSHL+ AL R ++TYID NL GD IS TLV+AI+++ +S+
Sbjct: 143 KKNDVFISFRGEDTRSNFTSHLHAALCRTKVKTYIDYNLKKGDYISETLVKAIQDSYVSI 322
Query: 78 IVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN 137
+VFS+NYA+S +CLDEL +++C + +VVP+FY+VDP+ VR Q GSY AF KH N
Sbjct: 323 VVFSENYASSTWCLDELTHMMKCLKNNQIVVVPVFYNVDPSHVRKQSGSYMVAFEKHVCN 502
Query: 138 SEEKIKVQEWR 148
KV +WR
Sbjct: 503 LNHFNKVNDWR 535
>BF648597 weakly similar to GP|12003378|gb Avr9/Cf-9 rapidly elicited protein
4 {Nicotiana tabacum}, partial (21%)
Length = 630
Score = 149 bits (377), Expect = 4e-37
Identities = 79/163 (48%), Positives = 109/163 (66%), Gaps = 6/163 (3%)
Frame = +1
Query: 5 YSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEIS 63
YSSS+SSS N PQ H+VFI+FR ED R F SHL L I T++DN L G++I
Sbjct: 142 YSSSSSSSTN-PQYLHDVFINFRGEDVRRTFVSHLYAVLSNAGINTFLDNEKLEKGEDIG 318
Query: 64 TTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQ 123
L++AI + +S+IVFSKNY S +CL+EL KI+EC+ + G +V+P+FYDVDP+ VR+Q
Sbjct: 319 HELLQAISVSRISIIVFSKNYTESSWCLNELEKIMECRXLHGHVVLPVFYDVDPSXVRHQ 498
Query: 124 RGSYAEAF---AKHEKNSEEKI--KVQEWRNGLMEAANYSGWD 161
+G + +A AK EE + ++ +WR L EA+N SGWD
Sbjct: 499 KGDFGKALEVAAKSRYIIEEVMGKELGKWRKVLTEASNLSGWD 627
>TC80590 similar to GP|23477201|emb|CAD36199. NLS-TIR-NBS disease resistance
protein {Populus tremula}, partial (13%)
Length = 679
Score = 148 bits (374), Expect = 9e-37
Identities = 68/104 (65%), Positives = 87/104 (83%), Gaps = 1/104 (0%)
Frame = +2
Query: 64 TTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQ 123
+ L+RAIEEA LSV+VFSKNY SK+CLDEL+KILECK+M+G++V+PIFYD++P+DVRNQ
Sbjct: 2 SALLRAIEEASLSVVVFSKNYGNSKWCLDELVKILECKKMRGQIVLPIFYDIEPSDVRNQ 181
Query: 124 RGSYAEAFAKHEKNSEEKI-KVQEWRNGLMEAANYSGWDCNVNR 166
GSYA+AF KHE+ + +VQ+WR L EAAN SGWDC+ NR
Sbjct: 182 TGSYADAFVKHEERFHGNLERVQKWREALREAANLSGWDCSTNR 313
>TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (22%)
Length = 832
Score = 148 bits (374), Expect = 9e-37
Identities = 77/163 (47%), Positives = 107/163 (65%), Gaps = 2/163 (1%)
Frame = +1
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDN-NLNSG 59
MA S SS++ ++ + ++VF++FR EDTRNNFT L AL+ I + D NL G
Sbjct: 31 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 210
Query: 60 DEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTD 119
+ I L RAIE +++ V +FSKNYA+S +CL EL KI EC + GK V+P+FYDVDP++
Sbjct: 211 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 390
Query: 120 VRNQRGSYAEAFAKHEKN-SEEKIKVQEWRNGLMEAANYSGWD 161
VR Q G Y+EAF KHE+ ++ +KV WR L + + SGWD
Sbjct: 391 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWD 519
>BI309457 weakly similar to GP|15787901|gb resistance gene analog NBS7
{Helianthus annuus}, partial (45%)
Length = 806
Score = 148 bits (374), Expect = 9e-37
Identities = 77/163 (47%), Positives = 109/163 (66%), Gaps = 2/163 (1%)
Frame = +3
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDN-NLNSG 59
MA + +SS+ ++ + ++VF++FR EDTRNNFT L AL+ I + D+ NL G
Sbjct: 15 MASTSNSSSVLGTSSRRNYYDVFVTFRGEDTRNNFTDFLFDALQTKGIIVFSDDTNLPKG 194
Query: 60 DEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTD 119
+ I L+RAIE +++ V VFS NYA+S +CL EL KI EC + GK V+P+FYDVDP+D
Sbjct: 195 ESIGPELLRAIEGSQVFVAVFSINYASSTWCLQELEKICECVKGSGKHVLPVFYDVDPSD 374
Query: 120 VRNQRGSYAEAFAKHEKNSEEKI-KVQEWRNGLMEAANYSGWD 161
VR Q G Y EAF KHE+ +++ KV +WR+ L + + SGWD
Sbjct: 375 VRKQSGIYGEAFIKHEQRFQQEFQKVSKWRDALKQVGSISGWD 503
>TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (12%)
Length = 632
Score = 147 bits (372), Expect = 1e-36
Identities = 73/149 (48%), Positives = 104/149 (68%), Gaps = 5/149 (3%)
Frame = +3
Query: 17 QEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAEL 75
Q VF+SFR DTRNNFT +L AL IRT+ID+N L GDEI+ +LV+AIEE+ +
Sbjct: 39 QSPSRVFLSFRGSDTRNNFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVKAIEESRI 218
Query: 76 SVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHE 135
+ +FS NYA+S FCLDEL+ I+ C + K +V+P+FYDV+PT +R+Q GSY E KHE
Sbjct: 219 FIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEHLTKHE 398
Query: 136 K----NSEEKIKVQEWRNGLMEAANYSGW 160
+ N + ++++W+ L++AAN SG+
Sbjct: 399 ERFQNNEKNMERLRQWKIALIQAANLSGY 485
>TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance protein
{Arabidopsis thaliana}, partial (7%)
Length = 1043
Score = 139 bits (350), Expect(2) = 3e-36
Identities = 66/131 (50%), Positives = 98/131 (74%), Gaps = 1/131 (0%)
Frame = +1
Query: 6 SSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEIST 64
SSS+ ++++ ++H VF++FR EDTR NFTSHL AL I T++D+ + GD IS
Sbjct: 193 SSSSFGASHSNSKQHHVFLNFRGEDTRYNFTSHLYAALCGKKIHTFMDDEEIERGDNISP 372
Query: 65 TLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQR 124
TL+ AIE +++ VI+FS++YA+S +CLDEL+KI+EC K +V+P+FY +D + VR+QR
Sbjct: 373 TLLSAIESSKICVIIFSQDYASSSWCLDELVKIIECSEKKKLVVIPVFYHIDASHVRHQR 552
Query: 125 GSYAEAFAKHE 135
G+Y +AFAKHE
Sbjct: 553 GTYGDAFAKHE 585
Score = 28.5 bits (62), Expect(2) = 3e-36
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = +2
Query: 143 KVQEWRNGLMEAANYSGWDCNVNR 166
KV WR L +AAN +GW NR
Sbjct: 611 KVHMWRTALHKAANLAGWVSEKNR 682
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,852,729
Number of Sequences: 36976
Number of extensions: 54801
Number of successful extensions: 507
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 392
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 396
length of query: 166
length of database: 9,014,727
effective HSP length: 89
effective length of query: 77
effective length of database: 5,723,863
effective search space: 440737451
effective search space used: 440737451
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC134049.18


