

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.1 - phase: 0
(630 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
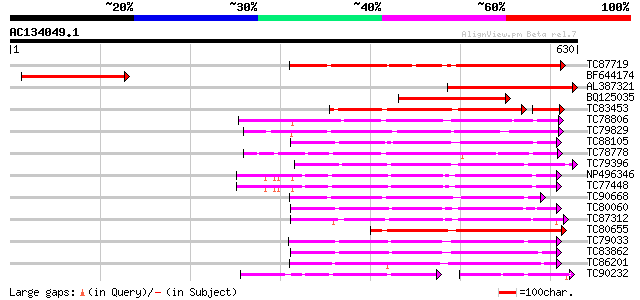
Score E
Sequences producing significant alignments: (bits) Value
TC87719 similar to PIR|E96692|E96692 probable wall-associated ki... 267 8e-72
BF644174 weakly similar to PIR|E96721|E967 hypothetical protein ... 261 6e-70
AL387321 similar to PIR|E96721|E96 hypothetical protein T17F3.6 ... 259 3e-69
BQ125035 similar to PIR|E96721|E96 hypothetical protein T17F3.6 ... 249 2e-66
TC83453 similar to GP|19715597|gb|AAL91622.1 At2g23450/F26B6.10 ... 194 2e-54
TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein ki... 208 6e-54
TC79829 similar to GP|20146232|dbj|BAB89014. similar to receptor... 205 4e-53
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 202 3e-52
TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-spe... 199 3e-51
TC79396 similar to GP|13605881|gb|AAK32926.1 At2g11520/F14P14.15... 198 5e-51
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 197 1e-50
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 197 1e-50
TC90668 similar to PIR|H86295|H86295 hypothetical protein AAF185... 196 2e-50
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 196 2e-50
TC87312 similar to GP|9759225|dbj|BAB09637.1 contains similarity... 195 4e-50
TC80655 similar to GP|19715597|gb|AAL91622.1 At2g23450/F26B6.10 ... 194 7e-50
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 189 2e-48
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 188 5e-48
TC86201 similar to PIR|T49003|T49003 protein kinase-like protein... 187 1e-47
TC90232 weakly similar to PIR|T45697|T45697 hypothetical protein... 131 3e-47
>TC87719 similar to PIR|E96692|E96692 probable wall-associated kinase
T4O24.5 [imported] - Arabidopsis thaliana, partial (32%)
Length = 1376
Score = 267 bits (683), Expect = 8e-72
Identities = 143/307 (46%), Positives = 205/307 (66%), Gaps = 1/307 (0%)
Frame = +1
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
VFTY+EL +TNNF +++G+GGFG+VY G+L+DG++ AVK RH + F N
Sbjct: 115 VFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVK---RHYESNFKRVAQFMN 285
Query: 372 EILILSSIDHPNLVKLHGYCSD-PRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTR 430
E+ IL+ + H NLV L+G S R L+LVY+Y+ NGT+A+HLHG +S ++ W R
Sbjct: 286 EVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSS--SCLLPWSVR 459
Query: 431 LEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTT 490
L+IA++TA A+ YLH S ++HRD+ S+NI +++ +KV DFGLSRL N T
Sbjct: 460 LDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLF----PNDVT 618
Query: 491 SSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMA 550
V T PQGTPGY+DP+Y++ ++LT+KSDVYSFGVVL+ELIS L+AVD R + ++
Sbjct: 619 H----VSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVN 786
Query: 551 LADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVG 610
LA+M V++I + +L +++DP L DN AVAELAFRC+ +D RP E+V
Sbjct: 787 LANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVE 966
Query: 611 ELKRVRS 617
L+ ++S
Sbjct: 967 VLRAIKS 987
>BF644174 weakly similar to PIR|E96721|E967 hypothetical protein T17F3.6
[imported] - Arabidopsis thaliana, partial (13%)
Length = 362
Score = 261 bits (667), Expect = 6e-70
Identities = 120/120 (100%), Positives = 120/120 (100%)
Frame = +1
Query: 14 LFPTSTHTCPTNNTTKTHPSPCPPFQSTPPFPFSTTPGCGHPSFQLTCSTPHSFITINNL 73
LFPTSTHTCPTNNTTKTHPSPCPPFQSTPPFPFSTTPGCGHPSFQLTCSTPHSFITINNL
Sbjct: 1 LFPTSTHTCPTNNTTKTHPSPCPPFQSTPPFPFSTTPGCGHPSFQLTCSTPHSFITINNL 180
Query: 74 TFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKPINLSNTPFTLSDETCSRLS 133
TFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKPINLSNTPFTLSDETCSRLS
Sbjct: 181 TFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKPINLSNTPFTLSDETCSRLS 360
>AL387321 similar to PIR|E96721|E96 hypothetical protein T17F3.6 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 503
Score = 259 bits (661), Expect = 3e-69
Identities = 134/146 (91%), Positives = 138/146 (93%), Gaps = 2/146 (1%)
Frame = +1
Query: 487 NQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDK 546
+QTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDK
Sbjct: 58 HQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDK 237
Query: 547 REMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSK 606
REMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSK
Sbjct: 238 REMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSK 417
Query: 607 EVVGELK-RVRSRI-SGGITRSMSTT 630
EVVG ++ RVRSR + RSMSTT
Sbjct: 418 EVVGAIEPRVRSRD*VVALLRSMSTT 495
>BQ125035 similar to PIR|E96721|E96 hypothetical protein T17F3.6 [imported] -
Arabidopsis thaliana, partial (18%)
Length = 392
Score = 249 bits (636), Expect = 2e-66
Identities = 124/124 (100%), Positives = 124/124 (100%)
Frame = +2
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS
Sbjct: 20 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 199
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA
Sbjct: 200 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 379
Query: 553 DMVV 556
DMVV
Sbjct: 380 DMVV 391
>TC83453 similar to GP|19715597|gb|AAL91622.1 At2g23450/F26B6.10
{Arabidopsis thaliana}, partial (33%)
Length = 1258
Score = 194 bits (494), Expect(2) = 2e-54
Identities = 102/219 (46%), Positives = 145/219 (65%)
Frame = +2
Query: 356 HRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHG 415
HR N S + NEI ++SS+ HPNLV+L G + ILVY+++ NGTL +HL
Sbjct: 5 HRDNE----SIEQVMNEIKLISSVSHPNLVRLLGCSIEYGEQILVYEFMSNGTLNQHLQ- 169
Query: 416 SKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDF 475
+ G ++W RL+IA +TA A+ +LH ++ PPI HRDI SSNI ++++ KV DF
Sbjct: 170 ---RETGNGLSWPVRLKIATETAQAISHLHSAIDPPIYHRDIKSSNILLDQNFGSKVADF 340
Query: 476 GLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELIS 535
GLSRL + + S+ + S T PQGTPGY+DP YH+ + L++KSDVYSFGVVL+E+I+
Sbjct: 341 GLSRLGITEISHASHVS-----TAPQGTPGYVDPQYHQDYHLSDKSDVYSFGVVLVEIIT 505
Query: 536 GLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDL 574
GLK VD+ R E+ LA + V RI G L +++DP++ L
Sbjct: 506 GLKVVDFSRPHNEVNLASLAVDRIRKGLLDDIIDPIIFL 622
Score = 37.0 bits (84), Expect(2) = 2e-54
Identities = 18/36 (50%), Positives = 25/36 (69%)
Frame = +3
Query: 581 LDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVR 616
L ++ VAELAFRC+A +D RP EV EL+++R
Sbjct: 654 LSSIHKVAELAFRCLAFHRDMRPCMTEVATELEQLR 761
>TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein kinase-like
{Arabidopsis thaliana}, partial (28%)
Length = 1962
Score = 208 bits (529), Expect = 6e-54
Identities = 134/366 (36%), Positives = 190/366 (51%), Gaps = 5/366 (1%)
Frame = +2
Query: 255 KPSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPV-- 312
K S V + ++L L+++I I R+ E V + N+ P +
Sbjct: 350 KSSHTGVIVGAVVAVLVLLVLAILIGIYAIRQKRARSSESNPFVNWEQNNNSGAAPQLKG 529
Query: 313 ---FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSF 369
F++DE+ TNNF IG GG+G VY G L G+L A+K + + A K+
Sbjct: 530 ARWFSFDEMRKYTNNFAEANTIGSGGYGQVYQGALPTGELVAIKRAGKESMQGAVEFKT- 706
Query: 370 CNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQT 429
EI +LS + H NLV L G+C + +LVY+YVPNGTL + L S++ G M W
Sbjct: 707 --EIELLSRVHHKNLVSLVGFCYEKGEQMLVYEYVPNGTLLDSL----SRKSGIWMDWIR 868
Query: 430 RLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQT 489
RL++ + A + YLH PPI+HRDI SSNI ++ + KV DFGLS+LLV
Sbjct: 869 RLKVTLGAARGLTYLHELADPPIIHRDIKSSNILLDNHLIAKVADFGLSKLLV------- 1027
Query: 490 TSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREM 549
S G V T +GT GYLDP+Y+ + +LTEKSDVYSFGV++LEL + K ++ +
Sbjct: 1028DSERGHVTTQVKGTMGYLDPEYYMTQQLTEKSDVYSFGVLMLELATSRKPIEQGKYIVRE 1207
Query: 550 ALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVV 609
+ M S+ L +LD L G L+ ELA RCV +RP EV
Sbjct: 1208VMRVMDTSK-ELYNLHSILDQSLLKGTRPKGLE---RYVELALRCVKEYAAERPSMAEVA 1375
Query: 610 GELKRV 615
E++ +
Sbjct: 1376KEIESI 1393
>TC79829 similar to GP|20146232|dbj|BAB89014. similar to receptor protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(40%)
Length = 1736
Score = 205 bits (522), Expect = 4e-53
Identities = 127/360 (35%), Positives = 194/360 (53%), Gaps = 4/360 (1%)
Frame = +2
Query: 260 AVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPP----VFTY 315
A+ IV+ + + LS ++++L R T +D AV H + + FTY
Sbjct: 191 AIVGIVLGAIACAVTLSAIVTLLILR----TKLKDYHAVSKRRHVSKIKIKMDGVRSFTY 358
Query: 316 DELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILI 375
+EL+ +TNNF ++G GG+G VY G + G A+K R + K F EI +
Sbjct: 359 EELSSATNNFSSSAQVGQGGYGKVYKGVISGGTAVAIK---RAQEGSLQGEKEFLTEISL 529
Query: 376 LSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAI 435
LS + H NLV L GYC + +LVY+Y+PNGTL +HL S + ++M RL+IA+
Sbjct: 530 LSRLHHRNLVSLIGYCDEEGEQMLVYEYMPNGTLRDHLSVSAKEPLTFIM----RLKIAL 697
Query: 436 QTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGF 495
+A + YLH PPI HRD+ +SNI ++ + KV DFGLSRL + + G
Sbjct: 698 GSAKGLMYLHNEADPPIFHRDVKASNILLDSKLSAKVADFGLSRLAPVPDMEGIVP--GH 871
Query: 496 VWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMV 555
V T +GTPGYLDP+Y + +LT+KSDVYS GVV LE+++G+ + + ++ +
Sbjct: 872 VSTVVKGTPGYLDPEYFLTHKLTDKSDVYSLGVVFLEILTGMHPISHGKN---------I 1024
Query: 556 VSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRV 615
V ++ V+ ++D + + V LA +CV + D+RP EVV EL+ +
Sbjct: 1025VREVNLSYQSGVIFSIIDERMGSYPSEHVEKFLTLALKCVNDEPDNRPTMAEVVRELENI 1204
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 202 bits (514), Expect = 3e-52
Identities = 121/301 (40%), Positives = 171/301 (56%)
Frame = +2
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
FT +L ++TN F IG+GG+G VY G L +G A+K L + A K F E
Sbjct: 689 FTLRDLELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKLLNNLGQA---EKEFRVE 859
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ + + H NLV+L G+C + +L+Y+YV NG L + LHG+ ++ GY+ TW R++
Sbjct: 860 VEAIGHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLHGAM-RQYGYL-TWDARIK 1033
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
I + TA A+ YLH +++P +VHRDI SSNI I+ D K+ DFGL++LL +S+ TT
Sbjct: 1034ILLGTAKALAYLHEAIEPKVVHRDIKSSNILIDDDFNAKISDFGLAKLLGAGKSHITTR- 1210
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
GT GY+ P+Y S L EKSDVYSFGV+LLE I+G VDY R E+ L
Sbjct: 1211-------VMGTFGYVAPEYANSGLLNEKSDVYSFGVLLLEAITGRDPVDYNRSAAEVNLV 1369
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
D + + +EV+DP ++ AL V A RCV D + RP +VV L
Sbjct: 1370DWLKMMVGNRHAEEVVDPNIETRPSTSALK---RVLLTALRCVDPDSEKRPKMSQVVRML 1540
Query: 613 K 613
+
Sbjct: 1541E 1543
>TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-specific
receptor protein kinase (EC 2.7.1.-) - Arabidopsis
thaliana, partial (23%)
Length = 1808
Score = 199 bits (505), Expect = 3e-51
Identities = 130/362 (35%), Positives = 201/362 (54%), Gaps = 7/362 (1%)
Frame = +3
Query: 260 AVFAIVIAFTSLILFLSVVISIL--RSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTYDE 317
A+ IV +S+ L L ++I+IL + R+ N E++ + + N +TY E
Sbjct: 543 AIVPIVATLSSVFLVL-LIIAILYWKLRRRNEPSEDE---MHMINKGTVVSKKLQYTYAE 710
Query: 318 LNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILS 377
+ T+N + IG GGFGSVY G ++DG AVK L + ++A K F E +L
Sbjct: 711 VLDITSNLEIV--IGRGGFGSVYSGQMKDGNKVAVKML---SASSAQGPKEFQTEAELLM 875
Query: 378 SIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQT 437
++ H NLV GYC + + L+Y+++ NG L E+L S++ + ++W+ RL+IAI
Sbjct: 876 TVHHKNLVSFIGYCDEGDKMALIYEFMANGNLKENL----SEKSSHCLSWERRLQIAIDA 1043
Query: 438 ALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVW 497
A ++YLH KPPI+HRD+ S+NI + +D+ K+ DFGLS++ + S+ V
Sbjct: 1044AEGLDYLHHGCKPPIIHRDVKSANILLNEDLEAKIADFGLSKVFKNSDIQNADSTLIHVD 1223
Query: 498 TGPQ-----GTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
+ GT GYLDP+Y++ L EKSD+YS+G+VLLELI+GL AV + K +
Sbjct: 1224VSGEKSAIMGTMGYLDPEYYKLQTLNEKSDIYSYGIVLLELITGLPAV--IKGKPSKHIL 1397
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
+ V R++ L +V+DP L+ D + + V LA C AS RP V+ EL
Sbjct: 1398EFVRPRLNKDDLSKVIDPRLEGKFD---VSSGWKVLGLAIACTASTSIQRPTMSVVLAEL 1568
Query: 613 KR 614
K+
Sbjct: 1569KQ 1574
>TC79396 similar to GP|13605881|gb|AAK32926.1 At2g11520/F14P14.15
{Arabidopsis thaliana}, partial (53%)
Length = 1658
Score = 198 bits (504), Expect = 5e-51
Identities = 117/314 (37%), Positives = 177/314 (56%)
Frame = +1
Query: 317 ELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILIL 376
+++ +T NF +IG+GGFG+VY +L DG + AVK R + + + F +E+ +L
Sbjct: 307 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTE--FSSEVELL 480
Query: 377 SSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQ 436
+ IDH NLVKL GY IL+ ++V NGTL EHL G + K ++ + RLEIAI
Sbjct: 481 AKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGK----ILDFNQRLEIAID 648
Query: 437 TALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFV 496
A + YLH + I+HRD+ SSNI + + MR KV DFG ++L + + S
Sbjct: 649 VAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHIS----- 813
Query: 497 WTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVV 556
T +GT GYLDP+Y +++ LT KSDVYSFG++LLE+++G + V+ + E
Sbjct: 814 -TKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAF 990
Query: 557 SRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVR 616
+ + G + +LDP++ + D + +LAF C A + DRPD K V +L +R
Sbjct: 991 RKYNEGSVVALLDPLM---QEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIR 1161
Query: 617 SRISGGITRSMSTT 630
+ +S STT
Sbjct: 1162A----DYLKSSSTT 1191
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 197 bits (500), Expect = 1e-50
Identities = 135/375 (36%), Positives = 208/375 (55%), Gaps = 14/375 (3%)
Frame = +1
Query: 253 KMKPSKIAVFAIVIAFTSLILFLSVVISIL---RSRKVNTTVE----EDPTA---VFLHN 302
K K K ++ A TS L +++ + IL R R + T+E P A +F
Sbjct: 1462 KAKKPKFGQVFVIGAITSGSLLITLAVGILFFCRYRHKSITLEGFGKTYPMATNIIFSLP 1641
Query: 303 HRNANLLPPV----FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRH 358
++ + V FT + + +T + K IG+GGFGSVY G L DG+ AVK
Sbjct: 1642 SKDDFFIKSVSVKPFTLEYIEQATEQY--KTLIGEGGFGSVYRGTLDDGQEVAVKV---R 1806
Query: 359 NHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKS 418
+ T+ ++ F NE+ +LS+I H NLV L GYC++ ILVY ++ NG+L + L+G S
Sbjct: 1807 SSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRLYGEAS 1986
Query: 419 KRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLS 478
KRK ++ W TRL IA+ A + YLH ++HRD+ SSNI +++ M KV DFG S
Sbjct: 1987 KRK--ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCAKVADFGFS 2160
Query: 479 RLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLK 538
+ QE + +V +GT GYLDP+Y+++ +L+EKSDV+SFGVVLLE++SG +
Sbjct: 2161 KYAP-QEGDS------YVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGRE 2319
Query: 539 AVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASD 598
++ R + E +L + I ++ E++DP + G +AL V E+A +C+
Sbjct: 2320 PLNIKRPRIEWSLVEWAKPYIRASKVDEIVDPGIKGGYHAEAL---WRVVEVALQCLEPY 2490
Query: 599 KDDRPDSKEVVGELK 613
RP ++V EL+
Sbjct: 2491 STYRPCMVDIVRELE 2535
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 197 bits (500), Expect = 1e-50
Identities = 135/375 (36%), Positives = 208/375 (55%), Gaps = 14/375 (3%)
Frame = +1
Query: 253 KMKPSKIAVFAIVIAFTSLILFLSVVISIL---RSRKVNTTVE----EDPTA---VFLHN 302
K K K ++ A TS L +++ + IL R R + T+E P A +F
Sbjct: 1906 KAKKPKFGQVFVIGAITSGSLLITLAVGILFFCRYRHKSITLEGFGKTYPMATNIIFSLP 2085
Query: 303 HRNANLLPPV----FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRH 358
++ + V FT + + +T + K IG+GGFGSVY G L DG+ AVK
Sbjct: 2086 SKDDFFIKSVSVKPFTLEYIEQATEQY--KTLIGEGGFGSVYRGTLDDGQEVAVKV---R 2250
Query: 359 NHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKS 418
+ T+ ++ F NE+ +LS+I H NLV L GYC++ ILVY ++ NG+L + L+G S
Sbjct: 2251 SSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRLYGEAS 2430
Query: 419 KRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLS 478
KRK ++ W TRL IA+ A + YLH ++HRD+ SSNI +++ M KV DFG S
Sbjct: 2431 KRK--ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCAKVADFGFS 2604
Query: 479 RLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLK 538
+ QE + +V +GT GYLDP+Y+++ +L+EKSDV+SFGVVLLE++SG +
Sbjct: 2605 KYAP-QEGDS------YVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGRE 2763
Query: 539 AVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASD 598
++ R + E +L + I ++ E++DP + G +AL V E+A +C+
Sbjct: 2764 PLNIKRPRIEWSLVEWAKPYIRASKVDEIVDPGIKGGYHAEAL---WRVVEVALQCLEPY 2934
Query: 599 KDDRPDSKEVVGELK 613
RP ++V EL+
Sbjct: 2935 STYRPCMVDIVRELE 2979
>TC90668 similar to PIR|H86295|H86295 hypothetical protein AAF18507.1
[imported] - Arabidopsis thaliana, partial (27%)
Length = 1243
Score = 196 bits (499), Expect = 2e-50
Identities = 109/284 (38%), Positives = 168/284 (58%)
Frame = +2
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
+FT +EL +T+N++ R +G GG+G+VY G L DG + AVK + ++F N
Sbjct: 437 LFTAEELQRATDNYNRSRFLGQGGYGTVYKGMLPDGTIVAVK---KSKELERNQIETFVN 607
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
E++ILS I+H N+VKL C + +LVY+++PNGTL++H+H K + ++W+ RL
Sbjct: 608 EVVILSQINHRNIVKLLSCCLETETPLLVYEFIPNGTLSQHIH---MKDQESSLSWENRL 778
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
IA + A A+ Y+HFS PI HRDI +NI ++ + KV DFG SR + L +++ TT
Sbjct: 779 RIACEVAGAVAYMHFSASIPIFHRDIKPTNILLDSNFSAKVSDFGTSRSIPLDKTHLTTF 958
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMAL 551
G GT GY+DP+Y +S + T KSDVYSFGVVL+ELI+ K + + + L
Sbjct: 959 VG--------GTYGYIDPEYFQSNQFTNKSDVYSFGVVLVELITSRKPISFYDEDDGQNL 1114
Query: 552 ADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCV 595
+S + Q+ +++D L D + A++ LA RC+
Sbjct: 1115IAHFISVMKENQVSQIIDARL---QKEAGKDTILAISSLARRCL 1237
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 196 bits (499), Expect = 2e-50
Identities = 115/301 (38%), Positives = 173/301 (57%)
Frame = +1
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
++ ++ ++TNNFDPK KIG+GGFG VY G L DG + AVK L + ++ F NE
Sbjct: 157 YSLRQIKVATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQG---NREFVNE 327
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
I ++S++ HPNLVKL+G C + L+LVY+Y+ N +LA L G +R + W+TR++
Sbjct: 328 IGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGKPEQRLN--LDWRTRMK 501
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
I + A + YLH + IVHRDI ++N+ ++K++ K+ DFGL++L E T S
Sbjct: 502 ICVGIARGLAYLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAKL---DEEENTHIS 672
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
T GT GY+ P+Y LT+K+DVYSFGVV LE++SG+ +Y + + L
Sbjct: 673 -----TRIAGTIGYMAPEYAMRGYLTDKADVYSFGVVALEIVSGMSNTNYRPKEEFVYLL 837
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
D G L E++DP LG+ + +A+ + +LA C RP VV L
Sbjct: 838 DWAYVLQEQGNLLELVDPT--LGSKYSSEEAM-RMLQLALLCTNPSPTLRPPMSSVVSML 1008
Query: 613 K 613
+
Sbjct: 1009E 1011
>TC87312 similar to GP|9759225|dbj|BAB09637.1 contains similarity to protein
kinase~gene_id:K19M22.13 {Arabidopsis thaliana}, partial
(43%)
Length = 1531
Score = 195 bits (496), Expect = 4e-50
Identities = 130/315 (41%), Positives = 176/315 (55%), Gaps = 7/315 (2%)
Frame = +3
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRH---NHTAAFSSKSF 369
F+++EL +T F P KIG+G FG VY G L DG L AVK + H A F
Sbjct: 363 FSFEELYKATGKFSPDNKIGEGAFGIVYKGRLYDGSLVAVKCARKDVQKKHLAEFK---- 530
Query: 370 CNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQT 429
NEI LS I+H NLV+ +GY I+V +YV NGTL EHL G + G +
Sbjct: 531 -NEINTLSKIEHLNLVRWYGYLEHGDDKIIVIEYVNNGTLREHLDGVR----GNGLEISE 695
Query: 430 RLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQT 489
RL+IAI A A+ YLH PI+HRDI +SNI I +R KV DFG +RL ++ N T
Sbjct: 696 RLDIAIDVAHAITYLHMYTDHPIIHRDIKASNILITDSLRAKVADFGFARLAP-EDPNAT 872
Query: 490 TSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREM 549
S T +GT GYLDPDY R+ +L+EKSDVYSFGV+L+E+++G V+ + E
Sbjct: 873 HIS-----TQVKGTAGYLDPDYMRTRQLSEKSDVYSFGVLLVEIMTGRHPVEPKKPLNER 1037
Query: 550 ALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSK--- 606
+ + G+ +DP L + ++ AV V +LAF+C+A + RP K
Sbjct: 1038VTIKWAMQLLKQGEAVIAMDPRLRRSSASN--KAVQKVLKLAFQCLAPVRRLRPSMKNCQ 1211
Query: 607 EVVGEL-KRVRSRIS 620
EV+ E+ K R R+S
Sbjct: 1212EVLWEIRKEFRDRVS 1256
>TC80655 similar to GP|19715597|gb|AAL91622.1 At2g23450/F26B6.10
{Arabidopsis thaliana}, partial (28%)
Length = 1126
Score = 194 bits (494), Expect = 7e-50
Identities = 101/217 (46%), Positives = 143/217 (65%)
Frame = +1
Query: 402 DYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSN 461
+Y+P TL++HL + +G + W RL IA +TA A+ YLH ++ PPI HRDI SSN
Sbjct: 1 EYMPQETLSQHLQ----RERGKGLPWTIRLTIASETANAIAYLHSAIHPPIYHRDIKSSN 168
Query: 462 IFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKS 521
I ++ + + KV DFGLSRL +++ S+ + T PQGTPGY+DP YH++F L++KS
Sbjct: 169 ILLDYNYKSKVADFGLSRLGLMETSH--------ISTAPQGTPGYVDPQYHQNFHLSDKS 324
Query: 522 DVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDAL 581
DVYSFGVVL+E+I+ +K VD+ R + E+ LA + V RI G + E++DP L+ D L
Sbjct: 325 DVYSFGVVLVEIITAMKVVDFGRPQSEINLAALAVDRIRRGSVDEIVDPFLEPNRDAWTL 504
Query: 582 DAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRSR 618
++ VAELAFRC+A D RP EV EL+ +R R
Sbjct: 505 YSIHKVAELAFRCLAFHSDTRPTMMEVAEELEYIRRR 615
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 189 bits (481), Expect = 2e-48
Identities = 113/306 (36%), Positives = 166/306 (53%), Gaps = 2/306 (0%)
Frame = +1
Query: 310 PP--VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSK 367
PP VF+ EL+ +TNNF+ K+G+GGFGSVY G L DG AVK L ++ A
Sbjct: 412 PPWRVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKA---DM 582
Query: 368 SFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTW 427
F E+ IL+ + H NL+ L GYC++ + ++VYDY+PN +L HLHG S ++ W
Sbjct: 583 EFAVEVEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSTES--LLDW 756
Query: 428 QTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESN 487
R+ IAI +A + YLH P I+HRD+ +SN+ ++ D + +V DFG ++L+
Sbjct: 757 NRRMNIAIGSAEGIVYLHVQATPHIIHRDVKASNVLLDSDFQARVADFGFAKLI------ 918
Query: 488 QTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKR 547
V T +GT GYL P+Y + E DVYSFG++LLEL SG K ++
Sbjct: 919 --PDGATHVTTRVKGTLGYLAPEYAMLGKANESCDVYSFGILLLELASGKKPLEKLSSSV 1092
Query: 548 EMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKE 607
+ A+ D + + E+ DP L N + + + V +A C + + RP E
Sbjct: 1093KRAINDWALPLACEKKFSELADPRL---NGDYVEEELKRVILVALICAQNQPEKRPTMVE 1263
Query: 608 VVGELK 613
VV LK
Sbjct: 1264VVELLK 1281
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 188 bits (478), Expect = 5e-48
Identities = 110/301 (36%), Positives = 167/301 (54%)
Frame = +1
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
++ EL +T+ F IG+ G+G VY G L+DG + AVK+L + A K F E
Sbjct: 13 YSLKELENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNLLNNKGQA---EKEFKVE 183
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ + + H NLV L GYC++ +LVY+YV NG L + LHG +TW R++
Sbjct: 184 VEAIGKVRHKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHGDVGPVSP--LTWDIRMK 357
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
IA+ TA + YLH ++P +VHRD+ SSNI ++K KV DFGL++LL S
Sbjct: 358 IAVGTAKGLAYLHEGLEPKVVHRDVKSSNILLDKKWHAKVSDFGLAKLL--------GSG 513
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
+V T GT GY+ P+Y + L E SDVYSFG++L+EL++G +DY R EM L
Sbjct: 514 KSYVTTRVMGTFGYVSPEYASTGMLNEGSDVYSFGILLMELVTGRSPIDYSRAPAEMNLV 693
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
D + + + +E++DP++++ +L V RC+ D + RP ++V L
Sbjct: 694 DWFKGMVASRRGEELVDPLIEIQPSPRSLKRALLV---CLRCIDLDANKRPKMGQIVHML 864
Query: 613 K 613
+
Sbjct: 865 E 867
>TC86201 similar to PIR|T49003|T49003 protein kinase-like protein -
Arabidopsis thaliana, partial (86%)
Length = 2127
Score = 187 bits (474), Expect = 1e-47
Identities = 115/307 (37%), Positives = 165/307 (53%), Gaps = 4/307 (1%)
Frame = +3
Query: 311 PVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFC 370
P + DEL T+NF K IG+G +G VY L DGK AVK L T S+ F
Sbjct: 372 PALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGKAVAVKKLDVS--TEPESNNEFL 545
Query: 371 NEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKS---KRKGYMMTW 427
++ ++S + + N V+LHGYC + +L Y++ G+L + LHG K + G + W
Sbjct: 546 TQVSMVSRLKNENFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLNW 725
Query: 428 QTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESN 487
R+ IA+ A +EYLH V+P I+HRDI SSN+ I +D + KV DF LS N
Sbjct: 726 MQRVRIAVDAARGLEYLHEKVQPSIIHRDIRSSNVLIFEDYKAKVADFNLS--------N 881
Query: 488 QTTSSGGFVW-TGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDK 546
Q + T GT GY P+Y + +LT+KSDVYSFGVVLLEL++G K VD+ +
Sbjct: 882 QAPDMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPR 1061
Query: 547 REMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSK 606
+ +L R+ ++K+ +DP L + + AVA L CV + + RP+
Sbjct: 1062GQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAAL---CVQYEAEFRPNMS 1232
Query: 607 EVVGELK 613
VV L+
Sbjct: 1233IVVKALQ 1253
>TC90232 weakly similar to PIR|T45697|T45697 hypothetical protein F18L15.120
- Arabidopsis thaliana, partial (26%)
Length = 1272
Score = 131 bits (329), Expect(2) = 3e-47
Identities = 79/223 (35%), Positives = 119/223 (52%)
Frame = +3
Query: 257 SKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTYD 316
+K V I+ + + L++ L + ++ R R+ L + A F+Y
Sbjct: 153 NKNIVVPIIGSLSGLVVILLISLAFWRFRRQKVGHSNSKKRGSLESTHEA------FSYT 314
Query: 317 ELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILIL 376
E+ TNNF K IG+GGFG VYLG L++ AVK L + ++ K F +E +L
Sbjct: 315 EILNITNNF--KTTIGEGGFGKVYLGILQNKTQVAVKML---SPSSMQGYKEFQSEAQLL 479
Query: 377 SSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQ 436
+ + H NLV L GYC + L+Y+Y+ NG L +HL S ++ W RL IA+
Sbjct: 480 AIVHHRNLVSLIGYCDEGEIKALIYEYMANGNLQQHLFVENSN----ILNWNERLNIAVD 647
Query: 437 TALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSR 479
A ++Y+H KPPI+HRD+ SNI ++ +M K+ DFGLSR
Sbjct: 648 AAQGLDYMHNGCKPPILHRDLKPSNILLDDNMHAKISDFGLSR 776
Score = 76.3 bits (186), Expect(2) = 3e-47
Identities = 46/133 (34%), Positives = 74/133 (55%), Gaps = 5/133 (3%)
Frame = +1
Query: 500 PQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRI 559
P GT GY DP+Y R+ +K+D+YSFG++L ELI+G KA+ + + + + V+ +
Sbjct: 817 PAGTLGYADPEYQRTGNTNKKNDIYSFGIILFELITGQKALTKASGE-NLHILEWVIPIV 993
Query: 560 HTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRS-- 617
G ++ V+D L ++++ V E+A C + D +RPD E++ ELK S
Sbjct: 994 EGGDIQNVVDSRL---QGEFSINSAWKVVEIAMSCTSPDVVERPDMSEILVELKECLSLD 1164
Query: 618 ---RISGGITRSM 627
R GG TR+M
Sbjct: 1165MVQRNCGG-TRAM 1200
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,657,268
Number of Sequences: 36976
Number of extensions: 429648
Number of successful extensions: 5405
Number of sequences better than 10.0: 724
Number of HSP's better than 10.0 without gapping: 4469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4695
length of query: 630
length of database: 9,014,727
effective HSP length: 102
effective length of query: 528
effective length of database: 5,243,175
effective search space: 2768396400
effective search space used: 2768396400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC134049.1


