

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133864.8 - phase: 0
(509 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
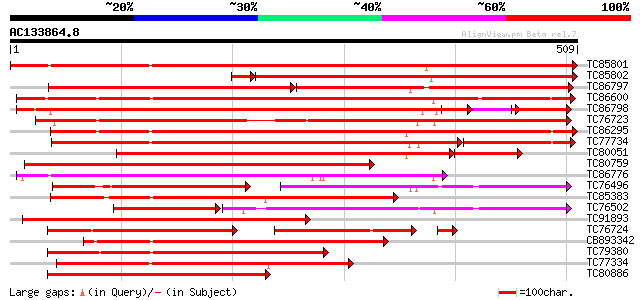
Score E
Sequences producing significant alignments: (bits) Value
TC85801 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-G... 847 0.0
TC85802 weakly similar to PIR|T47837|T47837 beta-glucosidase-lik... 580 e-174
TC86797 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosid... 293 e-146
TC86600 similar to GP|17226270|gb|AAL37714.1 beta-mannosidase en... 482 e-136
TC86798 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosid... 460 e-130
TC76723 beta glucosidase-like protein [Medicago truncatula] 440 e-124
TC86295 weakly similar to PIR|T02127|T02127 beta-glucosidase hom... 426 e-119
TC77734 weakly similar to PIR|T02128|T02128 beta-glucosidase hom... 342 e-116
TC80051 similar to GP|15778634|gb|AAL07489.1 amygdalin hydrolase... 351 e-109
TC80759 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-G... 382 e-106
TC86776 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosid... 340 8e-94
TC76496 weakly similar to PIR|S78099|S78099 furostanol glycoside... 195 5e-93
TC85383 similar to GP|19569603|gb|AAL92115.1 hydroxyisourate hyd... 335 2e-92
TC76502 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-G... 239 2e-88
TC91893 similar to SP|P26204|BGLS_TRIRP Non-cyanogenic beta-gluc... 316 1e-86
TC76724 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-G... 200 2e-86
CB893342 weakly similar to PIR|T49117|T49 glucosidase like prote... 294 6e-80
TC79380 similar to PIR|F86392|F86392 hypothetical protein AAF985... 285 2e-77
TC77334 weakly similar to PIR|T02128|T02128 beta-glucosidase hom... 283 1e-76
TC80886 similar to GP|15778431|gb|AAL07435.1 prunasin hydrolase ... 275 4e-74
>TC85801 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase
Mol_id: 1; Molecule: Cyanogenic Beta-Glucosidase; Chain:
Null; Ec: 3.2., partial (72%)
Length = 1732
Score = 847 bits (2188), Expect = 0.0
Identities = 406/515 (78%), Positives = 442/515 (84%), Gaps = 6/515 (1%)
Frame = +1
Query: 1 MKAISHFLLYLFSLATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEG 60
M AI LLYLFSLATLLAVVTGT + V PS YA SFNR+LFP DFLFGIGSSAYQ+EG
Sbjct: 16 MGAIGPSLLYLFSLATLLAVVTGTEPKKVEPSQYA-SFNRSLFPDDFLFGIGSSAYQVEG 192
Query: 61 ASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISW 120
AS+IDGRGPSIWDTFTKQ+P KI DHSSGN+GADFYHRYK D+K++KEIGLDSYRFSISW
Sbjct: 193 ASDIDGRGPSIWDTFTKQNPNKIWDHSSGNVGADFYHRYKEDLKVVKEIGLDSYRFSISW 372
Query: 121 SRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKI 180
SRIFP KGKGAVNP+GVKFYNN+INE+LANGL+PFVTLFHWDLPQ+LEDEYKGFL+ I
Sbjct: 373 SRIFP--KGKGAVNPLGVKFYNNLINEILANGLVPFVTLFHWDLPQALEDEYKGFLNKNI 546
Query: 181 VKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTT 240
KDF YADFCFKTFGDRVKHWVTLNEP SYTINGY+GGT P RCSKYV NC+TGDS+T
Sbjct: 547 AKDFAVYADFCFKTFGDRVKHWVTLNEPYSYTINGYNGGTFAPGRCSKYVANCTTGDSST 726
Query: 241 EPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDF 300
EPYIVAH+ ILSHAAA ++YK KYQAHQKGKIG+TL+TH++EPYSNSVAD KAA RALDF
Sbjct: 727 EPYIVAHNLILSHAAAVRVYKRKYQAHQKGKIGVTLVTHFFEPYSNSVADKKAAGRALDF 906
Query: 301 LFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPT 360
FGWFAHPITYGHYPQSMIS LG RLPKFTK E IIK SYDFLGVNYY+TYYAQS P
Sbjct: 907 FFGWFAHPITYGHYPQSMISLLGKRLPKFTKFESVIIKNSYDFLGVNYYSTYYAQSKGPQ 1086
Query: 361 YINMTYFTDMQA------NLIHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITEN 414
NM Y++DMQA N + WL VYPKGIH LVTHIK+ Y NPPVYITEN
Sbjct: 1087NKNMDYYSDMQATVSPLKNGVSIGPSTDLTWLYVYPKGIHDLVTHIKNVYNNPPVYITEN 1266
Query: 415 GIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGY 474
GI RNDS+P+NVARKDG+RI+YH HL +LL+ IK GANVKGYYAWSFSDSYEWDAGY
Sbjct: 1267GIATYRNDSVPINVARKDGVRIKYHHDHLYYLLEGIKHGANVKGYYAWSFSDSYEWDAGY 1446
Query: 475 TVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLKGKH 509
TVRFGIIYVDFVN LKRYPKYSAFWLQKFLLKGKH
Sbjct: 1447TVRFGIIYVDFVNKLKRYPKYSAFWLQKFLLKGKH 1551
>TC85802 weakly similar to PIR|T47837|T47837 beta-glucosidase-like protein -
Arabidopsis thaliana, partial (43%)
Length = 1119
Score = 580 bits (1494), Expect(2) = e-174
Identities = 279/295 (94%), Positives = 279/295 (94%), Gaps = 6/295 (2%)
Frame = +3
Query: 221 SPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHY 280
SPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHY
Sbjct: 66 SPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHY 245
Query: 281 YEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGS 340
YEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGS
Sbjct: 246 YEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGS 425
Query: 341 YDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIHS------NEEWSNYWLLVYPKGIH 394
YDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLI WL VYPKGIH
Sbjct: 426 YDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIPMKNGVTIGSSTDLNWLYVYPKGIH 605
Query: 395 HLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGA 454
HLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGA
Sbjct: 606 HLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGA 785
Query: 455 NVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLKGKH 509
NVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLKGKH
Sbjct: 786 NVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLKGKH 950
Score = 52.0 bits (123), Expect(2) = e-174
Identities = 21/21 (100%), Positives = 21/21 (100%)
Frame = +2
Query: 200 KHWVTLNEPVSYTINGYHGGT 220
KHWVTLNEPVSYTINGYHGGT
Sbjct: 2 KHWVTLNEPVSYTINGYHGGT 64
>TC86797 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase Mol_id:
1; Molecule: Cyanogenic Beta-Glucosidase; Chain: Null;
Ec: 3.2., partial (97%)
Length = 1517
Score = 293 bits (751), Expect(2) = e-146
Identities = 132/222 (59%), Positives = 172/222 (77%), Gaps = 1/222 (0%)
Frame = +3
Query: 36 ASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADF 95
+ NR+ FP +F+FG SSA+Q EGA+ DG+GPSIWDTFT +HPEKI D +G++ D
Sbjct: 60 SDLNRSCFPPNFVFGTASSAFQYEGAAFEDGKGPSIWDTFTHEHPEKIRDGGNGDVADDA 239
Query: 96 YHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIP 155
YHRYK DI I+K++ +D+YRFSI+WSR+ P K G VN G+ +YNN+I+E+LANGL P
Sbjct: 240 YHRYKEDIGIIKDLNMDAYRFSIAWSRVLPKGKLSGGVNQEGINYYNNLIDELLANGLQP 419
Query: 156 FVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTING 215
+VTLFHWD+PQ+LEDEY GFLSP+IV DF+ YA+ C+K FGDRVKHW+TLNEP S ++N
Sbjct: 420 YVTLFHWDVPQALEDEYDGFLSPRIVDDFKDYAELCYKEFGDRVKHWITLNEPWSVSMNA 599
Query: 216 YHGGTSPPARCSKYVG-NCSTGDSTTEPYIVAHHFILSHAAA 256
Y G P RCS ++ NC+ GDS TEPY+ AH+ +L+HAAA
Sbjct: 600 YAYGKFAPGRCSDWLNLNCTGGDSGTEPYLTAHYQLLAHAAA 725
Score = 245 bits (625), Expect(2) = e-146
Identities = 118/260 (45%), Positives = 175/260 (66%), Gaps = 11/260 (4%)
Frame = +2
Query: 258 KLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQS 317
KLYK KYQA Q G IGITL++H+YEP S + AD AA R LDF+FGW+ PIT G+YP+S
Sbjct: 728 KLYKNKYQASQYGIIGITLLSHWYEPASQAKADVDAAQRGLDFMFGWYMDPITKGNYPKS 907
Query: 318 MISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIP-----------PTYINMTY 366
M S +G+RLPKF+K+E + +KGS+DFLG+NYY++ YA P + IN T+
Sbjct: 908 MRSLVGSRLPKFSKKESEELKGSFDFLGLNYYSSSYAADAPHLRDARPAIQTDSLINATF 1087
Query: 367 FTDMQANLIHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPV 426
D + ++ WL +YP+G+ +L+ ++K+ Y NP +YITENG + + + +
Sbjct: 1088HRDGKP----LGPMSASSWLCIYPRGLGNLLLYVKNKYNNPVIYITENGRDEFNDPKLSL 1255
Query: 427 NVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFV 486
+ D RI Y HL ++ AI++G NVKGY+AWS D++EW++G+++RFG+++VD+
Sbjct: 1256EESLLDTYRIDYFYRHLYYVQAAIREGVNVKGYFAWSLLDNHEWESGFSLRFGLVFVDYN 1435
Query: 487 NNLKRYPKYSAFWLQKFLLK 506
++LKR+PK SA W + FL K
Sbjct: 1436DHLKRHPKLSAHWFKNFLKK 1495
>TC86600 similar to GP|17226270|gb|AAL37714.1 beta-mannosidase enzyme
{Lycopersicon esculentum}, partial (92%)
Length = 1737
Score = 482 bits (1241), Expect = e-136
Identities = 245/510 (48%), Positives = 329/510 (64%), Gaps = 8/510 (1%)
Frame = +1
Query: 7 FLLYLFSLATLLAVVTG-TASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNID 65
+ ++ F L L++ V G T + VH +R +FP F+FG+ +SAYQ+EG ++ +
Sbjct: 73 YSMFFFFLVVLVSSVNGVTVPETVHLD--TGGLSRDVFPKGFVFGVATSAYQVEGMASKE 246
Query: 66 GRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFP 125
GRGPSIWD F K+ P + ++ +G + D YHRYK DI ++ ++ D YRFSISWSRIFP
Sbjct: 247 GRGPSIWDVFIKK-PGIVANNGTGEVSVDQYHRYKEDIDLIAKLNFDQYRFSISWSRIFP 423
Query: 126 SKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFE 185
+ G G VN GV +YN +I+ +L G+ P+ L+H+DLP +LE +Y G LS +VKDF
Sbjct: 424 N--GTGKVNWKGVAYYNRLIDYLLEKGITPYANLYHYDLPLALELKYNGLLSRNVVKDFA 597
Query: 186 AYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIV 245
YADFCFKTFGDRVK+W+T NEP GY G P RCSK GNC+ G+S TEPY V
Sbjct: 598 DYADFCFKTFGDRVKNWMTFNEPRVVAALGYDNGFFAPGRCSKEYGNCTAGNSGTEPYTV 777
Query: 246 AHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWF 305
AH+ ILSHAAAA+ Y+ KYQ QKG+IGI L +YEP + S AD+ AA RA DF GWF
Sbjct: 778 AHNLILSHAAAAQRYREKYQEKQKGRIGILLDFVWYEPLTRSKADNYAAQRARDFHIGWF 957
Query: 306 AHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMT 365
HP+ YG YP++M + +GNRLPKFTKEE K++KGS DF+G+N YTTYY +
Sbjct: 958 IHPLVYGEYPRTMQNIVGNRLPKFTKEEVKLVKGSIDFVGINQYTTYYMYDPHNQKPKVP 1137
Query: 366 -YFTDMQANLIHSNE------EWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQ 418
Y +D A ++ +YWL P G++ + +IK+ YKNP ++++ENG+
Sbjct: 1138GYQSDWNAGFAYAKNGVPVGPRAYSYWLYNVPWGLYKSIMYIKEHYKNPTMFLSENGMDD 1317
Query: 419 SRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRF 478
N + + D RI Y+ +L L +AI DGANV GY+AWS D++EW GYT RF
Sbjct: 1318PGN--VTFSKGLHDTTRINYYKGYLTQLKKAIDDGANVFGYHAWSLLDNFEWRLGYTSRF 1491
Query: 479 GIIYVDFVNNLKRYPKYSAFWLQKFLLKGK 508
GI+YVDF LKR PK SA+W ++ L K K
Sbjct: 1492GIVYVDF-KTLKRTPKMSAYWFKQLLTKKK 1578
>TC86798 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase Mol_id:
1; Molecule: Cyanogenic Beta-Glucosidase; Chain: Null;
Ec: 3.2., partial (97%)
Length = 1972
Score = 460 bits (1183), Expect = e-130
Identities = 224/419 (53%), Positives = 295/419 (69%), Gaps = 10/419 (2%)
Frame = +2
Query: 7 FLLYLFSLATLLAVVTGTASQHVHPSHYA--ASFNRTLFPSDFLFGIGSSAYQIEGASNI 64
F +F L +LL++V T + P H + FNRT FP F+FG SSA+Q EGA
Sbjct: 104 FNFNVFMLLSLLSIVV-THIDAIKPLHLQEFSDFNRTSFPPGFVFGTASSAFQYEGAVRE 280
Query: 65 DGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIF 124
G+GPSIWDTFT ++PEKI D +G++ D YHRYK DI IMK++ +D+YRFSISWSR+
Sbjct: 281 GGKGPSIWDTFTHKYPEKIRDRHNGDVADDSYHRYKEDIGIMKDLNMDAYRFSISWSRVL 460
Query: 125 PSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDF 184
P K G VN G+ +YN++INEVLA G+ P+VTLFHWD+PQ+LEDEY GFLS +IV DF
Sbjct: 461 PKGKFSGGVNQEGINYYNDLINEVLAKGMQPYVTLFHWDVPQALEDEYDGFLSRRIVDDF 640
Query: 185 EAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVG-NCSTGDSTTEPY 243
YA+ CFK FGDRVKHW+TLNEP S ++N Y G P RCS ++ NC+ GDS TEPY
Sbjct: 641 RDYAELCFKEFGDRVKHWITLNEPWSVSMNAYAYGKFAPGRCSDWLNLNCTGGDSGTEPY 820
Query: 244 IVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFG 303
+ AH+ +L+HAAA KLY+ KYQA Q GKIGITL++H+YEP S + +D AA R LDF+FG
Sbjct: 821 LAAHYQLLAHAAAVKLYRTKYQASQNGKIGITLLSHWYEPASQAKSDVDAALRGLDFMFG 1000
Query: 304 WFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIP-PTYI 362
W+ HPIT G+YP+SM S +GNRLP+F+K+E K +KGS+DFLG+NYY+++YA P P
Sbjct: 1001WYMHPITKGNYPKSMRSLVGNRLPRFSKKESKNLKGSFDFLGLNYYSSFYAADAPHPRNA 1180
Query: 363 NMTYFTD--MQANLIHSNEEW----SNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENG 415
TD + A H+ + ++ WL +YP+G L+ ++K Y +P +YITENG
Sbjct: 1181RPAIQTDSLINATFEHNGKPLGPMSASSWLCIYPRGFRQLLLYVKKHYNDPVIYITENG 1357
Score = 83.2 bits (204), Expect(2) = 3e-24
Identities = 31/54 (57%), Positives = 46/54 (84%)
Frame = +3
Query: 451 KDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFL 504
+DG NVKGY+AWS D++EW++G+++RFG+++VDF +NLKR+PK SA W + FL
Sbjct: 1656 RDGVNVKGYFAWSLLDNFEWESGFSLRFGLVFVDFKDNLKRHPKLSAHWFKNFL 1817
Score = 47.0 bits (110), Expect(2) = 3e-24
Identities = 23/71 (32%), Positives = 38/71 (53%)
Frame = +2
Query: 388 VYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLL 447
+YP+G L+ ++K Y +P +YITENG + + ++ + + D RI Y HL +L
Sbjct: 1466 IYPRGFRQLLLYVKKHYNDPVIYITENGRDEFNDPTLSLEESLLDTDRIDYFYRHLYYLQ 1645
Query: 448 QAIKDGANVKG 458
A + KG
Sbjct: 1646 TA*QGWRECKG 1678
>TC76723 beta glucosidase-like protein [Medicago truncatula]
Length = 2331
Score = 440 bits (1132), Expect = e-124
Identities = 231/500 (46%), Positives = 308/500 (61%), Gaps = 19/500 (3%)
Frame = +3
Query: 24 TASQHVHPSHYAASF--NRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPE 81
+ ++ V S YA SF NR+ FP F+FG GSS YQ EGA + DGRG WD F P
Sbjct: 216 SGAEPVIVSTYADSFELNRSSFPEGFVFGTGSSNYQYEGAVSEDGRGKGTWDIFAHT-PG 392
Query: 82 KIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFY 141
+ D + ++ D YHRYK D++IMK + D+YRFSISW RI P+ K VN G+ FY
Sbjct: 393 MVKDGKNADVAIDHYHRYKEDVQIMKNMNTDAYRFSISWPRIVPTGKISDGVNQAGIIFY 572
Query: 142 NNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKH 201
N+I E+LANG IP+VTLFHWDLPQ+L+D+Y GF+S I KDF+ + D CFK FGD VKH
Sbjct: 573 KNLIYELLANGQIPYVTLFHWDLPQALQDDYGGFVSENIRKDFKDFVDICFKEFGDSVKH 752
Query: 202 WVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYK 261
WVT NEP SYT+ ST++ Y H+ +L+HA +LYK
Sbjct: 753 WVTFNEPFSYTL------------------------STSDWYKSTHNQLLAHADVFELYK 860
Query: 262 AKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISS 321
YQA Q G IGI L +H+++PYS D KAA ALDF+FGWF P+T G YP S++S
Sbjct: 861 TTYQA-QNGVIGIGLNSHWFKPYSTDPLDQKAAEDALDFMFGWFIQPLTTGEYPASLVSY 1037
Query: 322 LGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYY-AQSIPPTYINM---------TYFTDMQ 371
+G++LPKFT E+ K + GSYDF+G+NYYT+ Y A + P I + F +
Sbjct: 1038VGDKLPKFTAEQSKSLIGSYDFIGINYYTSMYAANATKPIPIQSPSGGADGVNSVFKIVN 1217
Query: 372 ANLIHSNEE------WSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIP 425
L N++ W+ WL V PKGI L+ + K+ Y NP + ITENG+ + + ++
Sbjct: 1218VTLTDKNKDGTYIGAWAATWLYVCPKGIQDLLLYTKEKYNNPTIIITENGMNEVNDPTLS 1397
Query: 426 VNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDF 485
+ A D RI Y HL +LL A++ G V+GY+AWS D++EW+ GYTVRFGI +VD+
Sbjct: 1398LEEALMDTNRIDYFYRHLYYLLSAMRQGVKVQGYFAWSLLDNFEWNDGYTVRFGINFVDY 1577
Query: 486 VN-NLKRYPKYSAFWLQKFL 504
N +L R+PK SA W +KFL
Sbjct: 1578ENGHLTRHPKLSARWFRKFL 1637
>TC86295 weakly similar to PIR|T02127|T02127 beta-glucosidase homolog F8K4.2
- Arabidopsis thaliana, partial (80%)
Length = 1704
Score = 426 bits (1095), Expect = e-119
Identities = 218/483 (45%), Positives = 299/483 (61%), Gaps = 10/483 (2%)
Frame = +3
Query: 37 SFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFY 96
+ + +LFP +FLFG SS+YQ EGA DG+G + WD FT + I D ++G++ D Y
Sbjct: 96 NLSSSLFPRNFLFGTASSSYQFEGAFLSDGKGLNNWDVFTHK-TGTILDGTNGDVAVDHY 272
Query: 97 HRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGK-GAVNPMGVKFYNNVINEVLANGLIP 155
HRY+ D+ +M+ G++SYRFS+SW+RI P KG+ G VN G+ +YN +I+ ++ G+ P
Sbjct: 273 HRYQEDVDLMEHTGVNSYRFSLSWARILP--KGRFGKVNRAGINYYNRLIDALVDKGIEP 446
Query: 156 FVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTING 215
FVT+ H+D+PQ LE+ YK +LSP+I +DF YAD CFK FGDRVK+WVT NEP I G
Sbjct: 447 FVTITHYDIPQELEERYKSWLSPEIQEDFRYYADICFKYFGDRVKYWVTFNEPNVAVICG 626
Query: 216 YHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGIT 275
Y G PP+RCS GNCS G+S EP+I A + ILSH AA +Y+AKYQ +Q GKIGI
Sbjct: 627 YRTGLYPPSRCSDSFGNCSYGNSEREPFIAASNIILSHLAAVDVYRAKYQKNQGGKIGIA 806
Query: 276 LITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYK 335
+ +YEP+SNS D AA RA F WF PI G YP M LG L F+K + +
Sbjct: 807 MNAIWYEPFSNSTEDKLAAERAQSFYMNWFLDPIILGKYPAEMHEILGPDLLVFSKYDKE 986
Query: 336 IIKGSYDFLGVNYYTTYYA---------QSIPPTYINMTYFTDMQANLIHSNEEWSNYWL 386
K DF+G+N+YT+YY Q + T Q N E + W
Sbjct: 987 KFKNGLDFIGINHYTSYYVKDCIFSACEQGKGSSKTEGFALTSAQMNDKSIGEPTALAWF 1166
Query: 387 LVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFL 446
V+P+G+ ++VT+IKD Y N P++ITENG G S + D R+ Y S+L L
Sbjct: 1167YVHPQGMENIVTYIKDRYNNIPMFITENGFGTSESSYPTTEYELNDVKRVEYLSSYLDSL 1346
Query: 447 LQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLK 506
AI+ GA+VKGY+ WS D++EW+ GY++RFG+ +VDF L R P+ SAFW + F+ +
Sbjct: 1347ATAIRKGADVKGYFVWSILDNFEWNHGYSIRFGLHHVDFA-TLNRTPRGSAFWYKNFISE 1523
Query: 507 GKH 509
K+
Sbjct: 1524HKN 1532
>TC77734 weakly similar to PIR|T02128|T02128 beta-glucosidase homolog F8K4.3
- Arabidopsis thaliana, partial (73%)
Length = 1989
Score = 342 bits (878), Expect(2) = e-116
Identities = 177/379 (46%), Positives = 235/379 (61%), Gaps = 10/379 (2%)
Frame = +1
Query: 38 FNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYH 97
F+ + ++FLFG SSAYQ EGA DG+G S WD T + P+KI D ++G+I D YH
Sbjct: 97 FDPSPLSTNFLFGTASSAYQYEGAYLSDGKGLSDWDVVTHKTPDKIKDGTNGDIAVDQYH 276
Query: 98 RYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGK-GAVNPMGVKFYNNVINEVLANGLIPF 156
RY DI +M+ + ++SYRFSISW+R+ P KG+ G VN G+ +YN +I+ +L G+ PF
Sbjct: 277 RYLEDIDLMEAMKVNSYRFSISWARVLP--KGRFGEVNSGGISYYNRLIDALLLKGIEPF 450
Query: 157 VTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGY 216
VTL H+DLPQ L D Y+G+LSP+ +DF AD CFK+FGDRVK+W T NEP GY
Sbjct: 451 VTLSHFDLPQELGDRYEGWLSPESQEDFVYLADLCFKSFGDRVKYWATFNEPDYLITYGY 630
Query: 217 HGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITL 276
G +PP RCSK GNCS GDS EPY+ H+ ILSHAAAA +Y+ KYQA Q GKIGI L
Sbjct: 631 RKGIAPPFRCSKPFGNCSEGDSEKEPYLAVHNIILSHAAAAYIYRTKYQAEQGGKIGIVL 810
Query: 277 ITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKI 336
+YEP SNS+AD A RA F W PI +G YP M LG+ LPKF+ +
Sbjct: 811 HFDWYEPISNSMADKLATERARSFTNNWLLDPIIFGEYPPVMQKILGDILPKFSNNNKEK 990
Query: 337 IKGSYDFLGVNYYTTYYAQSI------PPTYINMT---YFTDMQANLIHSNEEWSNYWLL 387
+K DF+G+N+Y +YY + P I T + + + + + S W
Sbjct: 991 LKSGLDFIGINHYASYYIKDCIYSKCEPGPGITRTEGLFQQSAEKDGVPIGKPTSIDWQY 1170
Query: 388 VYPKGIHHLVTHIKDTYKN 406
VYP+G+ +VT++K Y N
Sbjct: 1171VYPQGMEKIVTYVKTRYNN 1227
Score = 93.2 bits (230), Expect(2) = e-116
Identities = 47/101 (46%), Positives = 65/101 (63%)
Frame = +2
Query: 408 PVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDS 467
P++ITENG G+ N + D R Y HL LL+AI+ GA+V+GY+AWS D+
Sbjct: 1232 PMFITENGYGELDNPNNTEEQYLNDFDRKNYMAGHLLSLLEAIRKGADVRGYFAWSLLDN 1411
Query: 468 YEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLKGK 508
+EW GYTVRFG+ +VD+ LKR P+ SA W ++F+ K K
Sbjct: 1412 FEWLQGYTVRFGLHHVDYA-TLKRTPRLSANWYKEFIAKHK 1531
>TC80051 similar to GP|15778634|gb|AAL07489.1 amygdalin hydrolase isoform AH
I precursor {Prunus serotina}, partial (66%)
Length = 1156
Score = 351 bits (900), Expect(2) = e-109
Identities = 168/311 (54%), Positives = 216/311 (69%), Gaps = 8/311 (2%)
Frame = +3
Query: 97 HRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPF 156
HRYK D++IMK++ +D+YRFSISWSRI P K VN G+ +YNN+INE+L GL PF
Sbjct: 39 HRYKEDVEIMKDMNMDAYRFSISWSRILPKGKASRGVNKEGINYYNNLINELLDKGLQPF 218
Query: 157 VTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGY 216
VTLFHWDLPQ+L++EY GFLSP IV DF YA+ C+K FGDRVKHW+TLNEP + + GY
Sbjct: 219 VTLFHWDLPQTLDEEYGGFLSPNIVNDFRDYAELCYKEFGDRVKHWITLNEPWTLSKYGY 398
Query: 217 HGGTSPPARCSKYVG-NCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGIT 275
G S P RCS + NC GDS TEPYIVAH+ +L+HA A K+YKAKYQA QKG IGIT
Sbjct: 399 ADGRSAPGRCSSWHDHNCIGGDSATEPYIVAHNQLLAHATAVKVYKAKYQASQKGSIGIT 578
Query: 276 LITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYK 335
L + P ++ +D +A RA+DF+ GWF P+T G YP SM S +G+RLPKF+K E K
Sbjct: 579 LSCDWMVPLHDTESDIRATERAVDFILGWFMEPLTTGDYPSSMQSLVGSRLPKFSKHEVK 758
Query: 336 IIKGSYDFLGVNYYTTYYA-------QSIPPTYINMTYFTDMQANLIHSNEEWSNYWLLV 388
++KGS+DF+G+NYYT+ YA +S P + T + N I S+ W+ +
Sbjct: 759 LVKGSFDFIGLNYYTSNYATDAPELSESRPSLLTDSQVITTSERNGIPIGPMTSSIWMSI 938
Query: 389 YPKGIHHLVTH 399
YPKGIH L+ +
Sbjct: 939 YPKGIHDLLLY 971
Score = 61.6 bits (148), Expect(2) = e-109
Identities = 29/61 (47%), Positives = 39/61 (63%)
Frame = +1
Query: 400 IKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGY 459
+K Y NP +YITENGI + + ++ + A D RI Y +HL +L AIKDG NVKGY
Sbjct: 973 LKTKYNNPLIYITENGIDELNDPTLSLEEALADTARIDYFYNHLYYLKSAIKDGVNVKGY 1152
Query: 460 Y 460
+
Sbjct: 1153 F 1155
>TC80759 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase
Mol_id: 1; Molecule: Cyanogenic Beta-Glucosidase; Chain:
Null; Ec: 3.2., partial (60%)
Length = 963
Score = 382 bits (981), Expect = e-106
Identities = 179/315 (56%), Positives = 231/315 (72%), Gaps = 1/315 (0%)
Frame = +2
Query: 14 LATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWD 73
L L +T + ++ V P +S NRT FP+ F+FG SS+YQ EGA+ GRG SIWD
Sbjct: 17 LFILTLFITLSFAEVVSPMLDVSSLNRTTFPTGFIFGTASSSYQYEGAAKEGGRGASIWD 196
Query: 74 TFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAV 133
T+T ++P+KI D S+G++ D Y+RYK D+ IM+ + LD+YRFSISWSRI P K KG +
Sbjct: 197 TYTHKYPDKIEDRSNGDVAVDQYYRYKEDVGIMRNMNLDAYRFSISWSRILPKGKLKGGI 376
Query: 134 NPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFK 193
N G+K+YNN+INE+L N L PFVTLFHWDLPQ+LEDEY GFLSP I+ DF+ YA+ CFK
Sbjct: 377 NQEGIKYYNNLINELLTNDLQPFVTLFHWDLPQALEDEYSGFLSPLIINDFQDYAELCFK 556
Query: 194 TFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYV-GNCSTGDSTTEPYIVAHHFILS 252
FGDRVK+W+T NEP SY+I GY G PP RCSK++ NC+ GDS EPYIV+HH +L+
Sbjct: 557 EFGDRVKYWITFNEPYSYSIGGYAIGFFPPGRCSKWLSSNCTDGDSGKEPYIVSHHQLLA 736
Query: 253 HAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYG 312
HAAA +YK KYQ QKG IGITL+++++ P+S++ D AA RA+DF+FGWF P+T G
Sbjct: 737 HAAAVDVYKKKYQESQKGVIGITLVSNWFIPFSDNKFDQNAAERAVDFMFGWFMEPLTTG 916
Query: 313 HYPQSMISSLGNRLP 327
YP+SM G RLP
Sbjct: 917 KYPKSMRFPGGKRLP 961
>TC86776 similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase Mol_id:
1; Molecule: Cyanogenic Beta-Glucosidase; Chain: Null;
Ec: 3.2., partial (65%)
Length = 1354
Score = 340 bits (872), Expect = 8e-94
Identities = 183/432 (42%), Positives = 255/432 (58%), Gaps = 45/432 (10%)
Frame = +2
Query: 7 FLLY--LFSLATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNI 64
+LL+ L SL + + G + P S NR FP F+FG SSAYQ EGA++
Sbjct: 47 YLLFSLLLSLVSTFIITEGKEIINTAPLQIG-SLNRNDFPEGFIFGTASSAYQYEGAASE 223
Query: 65 DGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIF 124
GRG SIWDTFT ++P+KI D ++G++ D YHRYK D+ IMK++ LD+YRFSISWSRI
Sbjct: 224 GGRGASIWDTFTHRYPQKITDGNNGDVAVDSYHRYKEDVGIMKDMNLDAYRFSISWSRIL 403
Query: 125 PSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDF 184
P K G +N G+ +YNN+I+E++ NGL PFVTLFHWDLPQ+LEDEY GFLSP I+KDF
Sbjct: 404 PKGKLSGGINQEGIDYYNNLIDELVTNGLQPFVTLFHWDLPQTLEDEYGGFLSPLIIKDF 583
Query: 185 EAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVG-NCSTGDSTTEPY 243
+ YA+ CFKTFGDRVKHW+TLNEP +Y+ +GY G P RCS ++ NC+ GDS TEPY
Sbjct: 584 QDYAELCFKTFGDRVKHWITLNEPWTYSQDGYANGEMAPGRCSSWLNPNCTGGDSGTEPY 763
Query: 244 IVAHHFILSHAAAAKLYKAKYQAHQKG----KIGITLI--------------THY----- 280
+VAH+ + S ++ + + + KG IG L T Y
Sbjct: 764 LVAHYQLTSSCSSC*FVQDQVSSLSKGCDRYNIGY*LFCATLR**T*YKGC*TSYRLYVW 943
Query: 281 ------------YEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPK 328
Y N++ + + ++ F P+ G YP++M + + +RLPK
Sbjct: 944 MVSSLQR*NLYNYTIQINTIKEIYMITYSISLFNYRFMDPLANGDYPKTMRALVRSRLPK 1123
Query: 329 FTKEEYKIIKGSYDFLGVNYYTTYYAQSIPP-TYINMTYFTDMQANLIHSNE------EW 381
FTKE+ K++ GS+DF+G+NYY++ YA P + +Y TD + +
Sbjct: 1124FTKEQSKLVSGSFDFIGINYYSSCYASDAPQLSNGKPSYLTDSLSRFSFERDGKTIGLNV 1303
Query: 382 SNYWLLVYPKGI 393
++ WL VYP+ I
Sbjct: 1304ASNWLYVYPRAI 1339
>TC76496 weakly similar to PIR|S78099|S78099 furostanol glycoside
26-O-beta-glucosidase F26G - Costus speciosus, partial
(35%)
Length = 1879
Score = 195 bits (495), Expect(2) = 5e-93
Identities = 101/271 (37%), Positives = 161/271 (59%), Gaps = 10/271 (3%)
Frame = +1
Query: 244 IVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFG 303
+V H+ ++SHA +KLYK+K+Q Q+G+IGI + + PYS+ D AA R ++F FG
Sbjct: 841 LVLHNLLISHATISKLYKSKFQTVQEGEIGIAISAKSFVPYSSKPQDVDAAQRLIEFQFG 1020
Query: 304 WFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIP----- 358
W P+ G YP+ M +G RLP+FTK E +++KGS DF+G+NYY + + + P
Sbjct: 1021 WVLEPLFKGDYPKIMRKLVGKRLPEFTKNEKEMLKGSTDFIGINYYFSLFVRHEPNRTKI 1200
Query: 359 PTYIN-----MTYFTDMQANLIHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITE 413
P N +T +++ N + +++ + VYP+G+++ + +I YKNP +YITE
Sbjct: 1201 PASDNFDALAVTEVLNVEGNTLGYYDQYGCSY--VYPEGLYNFLLYINKKYKNPRIYITE 1374
Query: 414 NGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAG 473
NGI S + KD R Y +H+ AI DG NV GY+AW+ D+Y++D G
Sbjct: 1375 NGI-----PSFNIPNPLKDEHRTAYIAAHINATKAAINDGLNVGGYFAWAAFDTYDFDDG 1539
Query: 474 YTVRFGIIYVDFVNNLKRYPKYSAFWLQKFL 504
Y+ G+ +++F ++LKR P +A W +K+L
Sbjct: 1540 YSKHMGLYHINFDDSLKRIPTKTAKWYKKYL 1632
Score = 164 bits (416), Expect(2) = 5e-93
Identities = 83/178 (46%), Positives = 112/178 (62%)
Frame = +3
Query: 39 NRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHR 98
NR FP F FG G+SA QIEG S+ GRG I D GD+ + Y R
Sbjct: 261 NRETFPRGFFFGAGTSAPQIEGGSHEGGRGLGILDVVYS------GDNKYVT-KIEHYQR 419
Query: 99 YKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVT 158
YK D++ +K +G++SYRFSISW+R+ P KG VN G++FYNN+INE+L N + PFVT
Sbjct: 420 YKEDVQRLKHLGVNSYRFSISWNRVIPDGTLKGGVNKEGIEFYNNLINELLNNDIEPFVT 599
Query: 159 LFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGY 216
+ H+D P +L+ GFL+ IVK F+ Y++ FKT+GDRVK+W T+NEP + Y
Sbjct: 600 ILHFDYPLALQQNIGGFLNRSIVKHFKDYSELLFKTYGDRVKYWTTMNEPELQAMYNY 773
>TC85383 similar to GP|19569603|gb|AAL92115.1 hydroxyisourate hydrolase
{Glycine max}, partial (59%)
Length = 1119
Score = 335 bits (860), Expect = 2e-92
Identities = 156/315 (49%), Positives = 220/315 (69%), Gaps = 2/315 (0%)
Frame = +3
Query: 37 SFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFY 96
+++R FP DF+FG G+SAYQ+EGA+N DGR PSIWDTF + G+ G++ D Y
Sbjct: 147 NYSRHDFPVDFVFGSGTSAYQVEGAANEDGRTPSIWDTFAHAGFARGGN---GDVACDTY 317
Query: 97 HRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPF 156
H+YK D+++M E GLD+YRFSISWSR+ P+ G+G VNP G+++YNN+INE++ NG+ P
Sbjct: 318 HKYKEDVQLMVETGLDAYRFSISWSRLIPN--GRGPVNPKGLQYYNNLINELIRNGIQPH 491
Query: 157 VTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGY 216
VTL ++DLPQ+LEDEY+G+LS +++KDF YAD CF+ FGDRVK+W T+NEP + + Y
Sbjct: 492 VTLHNYDLPQALEDEYEGWLSRQVIKDFTNYADVCFREFGDRVKYWTTVNEPNIFAVGSY 671
Query: 217 HGGTSPPARCSK--YVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGI 274
G SPP RCS V + G+ST EPY+V HH +L+H++A +LY+ KY+ Q G +GI
Sbjct: 672 DQGISPPKRCSPPFCVIESTKGNSTFEPYLVVHHILLAHSSAVRLYRRKYREEQNGFVGI 851
Query: 275 TLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEY 334
++ P +N+ D A R DF GW P+ +G YP SM ++ G R+P FT E
Sbjct: 852 SIYAFGSVPQTNTEKDRAACQRFHDFYLGWIMEPLLHGDYPDSMKANAGARIPSFTSRES 1031
Query: 335 KIIKGSYDFLGVNYY 349
+ +KGSYDF+G+ +Y
Sbjct: 1032EQVKGSYDFIGIIHY 1076
>TC76502 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase
Mol_id: 1; Molecule: Cyanogenic Beta-Glucosidase; Chain:
Null; Ec: 3.2., partial (32%)
Length = 1738
Score = 239 bits (610), Expect(2) = 2e-88
Identities = 135/324 (41%), Positives = 188/324 (57%), Gaps = 11/324 (3%)
Frame = +2
Query: 192 FKTFGDRVKHWVTLNEP---VSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHH 248
FKTFGDRVKHW TLNEP V Y G S + +C T TE Y H
Sbjct: 647 FKTFGDRVKHWATLNEPEVQVVYESVDNVGNWS--------MESCQTTKVCTEIYTELHI 802
Query: 249 FILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHP 308
+++HA A+KLYK+K+QA QKG+IGIT+ + Y PYS+ + D AA R DF +GW P
Sbjct: 803 LLIAHATASKLYKSKFQAIQKGEIGITISSESYVPYSSKLEDVDAAQRLTDFTWGWVLEP 982
Query: 309 ITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIP-PTYINMTYF 367
+ +G YPQ M +G RLPKFTK E +++KGS DF+G+NYY+++Y + P T + YF
Sbjct: 983 LFHGDYPQIMRKLVGKRLPKFTKNEKEMLKGSIDFIGINYYSSHYVRHEPNRTKVTGGYF 1162
Query: 368 TDMQANLIHSNEE------WSNY-WLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSR 420
D ANL N E W Y VYP+G+++ + ++ YKN +YI ENGI
Sbjct: 1163-DALANLEDINAEGKTLGYWDQYGGTYVYPEGLYNFLLYLNKKYKNSKIYINENGI---- 1327
Query: 421 NDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGI 480
SI + D R + +H+ AI DG NV+GY+AW+ D++++ GY+ G+
Sbjct: 1328-PSIKIPNPLNDEHRTAFIAAHINATKSAIDDGVNVRGYFAWAAFDTFDFYDGYSHNMGL 1504
Query: 481 IYVDFVNNLKRYPKYSAFWLQKFL 504
+VDF + LKR P +A W +K+L
Sbjct: 1505YHVDFNDCLKRIPTNTAKWYKKYL 1576
Score = 105 bits (262), Expect(2) = 2e-88
Identities = 45/96 (46%), Positives = 68/96 (69%)
Frame = +1
Query: 94 DFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGL 153
+ Y RYK D++ +K +G++SYR SI W+R+ P KG +N G+ FYNN+INE+L NG+
Sbjct: 352 EHYKRYKEDVQHLKNLGVNSYRMSICWNRVIPDGTLKGGINKEGINFYNNLINELLNNGI 531
Query: 154 IPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYAD 189
PFVT+ H D P +L+ ++ GFL+ IVK F+ Y++
Sbjct: 532 EPFVTILHLDYPLALQKKFGGFLNHSIVKHFKDYSE 639
>TC91893 similar to SP|P26204|BGLS_TRIRP Non-cyanogenic beta-glucosidase
precursor (EC 3.2.1.21). [Creeping white clover]
{Trifolium repens}, partial (46%)
Length = 788
Score = 316 bits (810), Expect = 1e-86
Identities = 151/260 (58%), Positives = 184/260 (70%), Gaps = 1/260 (0%)
Frame = +2
Query: 12 FSLATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSI 71
F+L A V + V P AS NR FP F+FG GSS+YQ EGA+N GR PSI
Sbjct: 8 FALGLGKANVNCIETIEVEPLLDVASLNRNSFPKGFIFGAGSSSYQFEGAANEGGRTPSI 187
Query: 72 WDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKG 131
WDTFT ++PEKI D S+ ++ D YH YK D++I+K++ LDSYRFS+SWSRI P K G
Sbjct: 188 WDTFTHKYPEKIKDRSNADVAIDSYHNYKEDVRIIKDMNLDSYRFSLSWSRILPKGKLSG 367
Query: 132 AVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFC 191
+N G+ +YNN INE+LANG+ P VTLFHWDLPQ+LEDEY GFLSP IVKDF+ YA+ C
Sbjct: 368 GINQEGINYYNNFINELLANGIKPLVTLFHWDLPQTLEDEYGGFLSPLIVKDFQDYAELC 547
Query: 192 FKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVG-NCSTGDSTTEPYIVAHHFI 250
FK FGDRVK+WVTLNEP SY+ NGY G P RCS ++ NC+ GDS EPYIV H+ +
Sbjct: 548 FKEFGDRVKYWVTLNEPWSYSQNGYANGRMAPGRCSSWLNPNCTGGDSAIEPYIVTHYQL 727
Query: 251 LSHAAAAKLYKAKYQAHQKG 270
L+HA A +YK KYQ Q G
Sbjct: 728 LAHAEAVNVYKTKYQETQNG 787
>TC76724 weakly similar to GP|1311386|pdb|1CBG| Cyanogenic Beta-Glucosidase
Mol_id: 1; Molecule: Cyanogenic Beta-Glucosidase; Chain:
Null; Ec: 3.2., partial (41%)
Length = 1304
Score = 200 bits (509), Expect(3) = 2e-86
Identities = 90/170 (52%), Positives = 119/170 (69%)
Frame = +1
Query: 35 AASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGAD 94
++ +R+ FP F+FG GSS YQ EGA + GRG WD P ++ D + +I D
Sbjct: 223 SSELSRSSFPEGFVFGTGSSNYQYEGAVSEGGRGKGTWD-IASHTPGRVKDGKNADIAID 399
Query: 95 FYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLI 154
YHRYK D+ IMK + D+YRFSISW RI P+ K G +N G++FYNN+I+E+LANG I
Sbjct: 400 HYHRYKEDVAIMKYMNTDAYRFSISWPRILPNGKLSGGINQEGIRFYNNLIDELLANGQI 579
Query: 155 PFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVT 204
P+VTLFHWDLP L++EY+GF SP I+ DF+ + + CF+ FGDRVKHWVT
Sbjct: 580 PYVTLFHWDLPNILQEEYEGFCSPYIINDFKDFVEICFQEFGDRVKHWVT 729
Score = 135 bits (339), Expect(3) = 2e-86
Identities = 65/128 (50%), Positives = 86/128 (66%)
Frame = +2
Query: 238 STTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRA 297
ST+ Y H+ +LSHAA +LYK KYQ Q G IGI L +H+++PYS D +A RA
Sbjct: 758 STSHRYKATHNQLLSHAAVVELYKTKYQDSQNGVIGIGLNSHWFKPYSTDPLDQQATERA 937
Query: 298 LDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSI 357
LDF+FGWF P+T G YP +M+S + + LPKFT+E+ K + GSYDF+G+NYYTT YA +
Sbjct: 938 LDFMFGWFIQPLTTGEYPANMVSFVKD-LPKFTEEQSKSLIGSYDFIGINYYTTMYAANA 1114
Query: 358 PPTYINMT 365
I T
Sbjct: 1115TEALILKT 1138
Score = 23.5 bits (49), Expect(3) = 2e-86
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +3
Query: 385 WLLVYPKGIHHLVTHIKD 402
WL V PKGI L+ + K+
Sbjct: 1251 WLYVCPKGIQDLLLYTKE 1304
>CB893342 weakly similar to PIR|T49117|T49 glucosidase like protein -
Arabidopsis thaliana, partial (51%)
Length = 821
Score = 294 bits (752), Expect = 6e-80
Identities = 139/276 (50%), Positives = 190/276 (68%), Gaps = 2/276 (0%)
Frame = +1
Query: 67 RGPSIWDTFTKQHPEKIGDHS-SGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFP 125
R SIWDTF H G + +G+I D YH+YK D+++M ++GLD+YRFSISWSR+ P
Sbjct: 1 RKASIWDTFA--HAGNGGLYKGNGDIACDQYHKYKDDVQLMSKMGLDAYRFSISWSRLIP 174
Query: 126 SKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFE 185
G G +NP G+++YNN+INE+ G+ P VTL HWDLPQ+LEDEY G++S +++KDF
Sbjct: 175 D--GNGPINPKGLQYYNNLINELTNQGIQPHVTLNHWDLPQALEDEYGGWVSRRVIKDFT 348
Query: 186 AYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKY-VGNCSTGDSTTEPYI 244
AYAD CF+ FGDRVKHW T+NE ++ GY G PP RCS + NCS G+S+TEPY+
Sbjct: 349 AYADVCFREFGDRVKHWTTVNEGNVCSMGGYDAGFLPPQRCSSSPIFNCSKGNSSTEPYL 528
Query: 245 VAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGW 304
V HH +L+HA+A +LY+ Y+ Q+G IG L+ + P +N+ D AA RA DF GW
Sbjct: 529 VTHHMLLAHASATRLYRKMYKVKQQGFIGFNLLVFGFVPLTNTSEDIIAAQRAQDFYLGW 708
Query: 305 FAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGS 340
F +P +G YP +M ++G+RLP FT E ++KGS
Sbjct: 709 FLNPFIFGEYPATMKKNVGSRLPFFTSREANMVKGS 816
>TC79380 similar to PIR|F86392|F86392 hypothetical protein AAF98564.1
[imported] - Arabidopsis thaliana, partial (49%)
Length = 978
Score = 285 bits (730), Expect = 2e-77
Identities = 133/253 (52%), Positives = 179/253 (70%), Gaps = 1/253 (0%)
Frame = +1
Query: 35 AASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGAD 94
+A +R FP F+FG SSA+Q EGA DGRGPS+WDTF+ K+ D S+ ++ D
Sbjct: 229 SAEISRANFPHGFIFGTASSAFQYEGAVKEDGRGPSVWDTFSHTFG-KVTDFSNADVAVD 405
Query: 95 FYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLI 154
YHRY+ DI++MK++G+D+YRFSISWSRI+P+ G GA+N G+ YN IN +LA G+
Sbjct: 406 QYHRYEEDIQLMKDLGMDAYRFSISWSRIYPN--GSGAINQAGIDHYNKFINALLAKGIE 579
Query: 155 PFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTIN 214
P+VTL+HWDLPQ+L+D+YKG+LS I+KDF YA+ CF+ FGDRVKHW+T NEP ++T
Sbjct: 580 PYVTLYHWDLPQALDDKYKGWLSTDIIKDFATYAETCFQKFGDRVKHWITFNEPHTFTTQ 759
Query: 215 GYHGGTSPPARCSKYVG-NCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIG 273
GY G P RCS + C G+S TEPYIVAH+ +L+HAA A +Y+ KY+ Q G +G
Sbjct: 760 GYDVGLQAPGRCSILLHLFCRAGNSATEPYIVAHNVLLTHAAVADIYRKKYKNTQGGSLG 939
Query: 274 ITLITHYYEPYSN 286
I +YEP +N
Sbjct: 940 IAFDVIWYEPATN 978
>TC77334 weakly similar to PIR|T02128|T02128 beta-glucosidase homolog F8K4.3
- Arabidopsis thaliana, partial (49%)
Length = 934
Score = 283 bits (724), Expect = 1e-76
Identities = 140/269 (52%), Positives = 178/269 (66%), Gaps = 3/269 (1%)
Frame = +1
Query: 43 FPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSD 102
FP++FLFG SS+YQ EGA N DG+G S WD FT I D S+G+I D YHRY+ D
Sbjct: 115 FPTNFLFGTASSSYQYEGAYNSDGKGQSNWDNFTHGGRGIIVDGSNGDIAVDHYHRYQED 294
Query: 103 IKIMKEIGLDSYRFSISWSRIFPSKKGK-GAVNPMGVKFYNNVINEVLANGLIPFVTLFH 161
I +++++ ++S+R SISW+RI P KG+ G VN G+ FYN +++ ++ G+ PFVTL H
Sbjct: 295 INLLEDLEVNSHRLSISWARILP--KGRFGEVNWAGIDFYNKLLDALMLKGIQPFVTLSH 468
Query: 162 WDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTS 221
+D+PQ LED Y G LSP+ DF YAD CFKTFGDRVK W+T NEP GY G
Sbjct: 469 YDIPQELEDRYGGLLSPQSQDDFAFYADLCFKTFGDRVKFWITFNEPNYLASLGYRSGLF 648
Query: 222 PPARCSKYVG--NCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITH 279
PP RCS + C+ GDS EP++ AH+ ILSHAAA +Y+ KYQA QKG+IGI +
Sbjct: 649 PPRRCSGSLAIVTCNEGDSEKEPFVAAHNIILSHAAAVDIYRTKYQAEQKGRIGIVISHE 828
Query: 280 YYEPYSNSVADHKAASRALDFLFGWFAHP 308
+YEP SNS AD AA RA F F W P
Sbjct: 829 WYEPMSNSNADKLAAERARSFTFNWILTP 915
>TC80886 similar to GP|15778431|gb|AAL07435.1 prunasin hydrolase isoform PH
A precursor {Prunus serotina}, partial (36%)
Length = 685
Score = 275 bits (702), Expect = 4e-74
Identities = 123/200 (61%), Positives = 154/200 (76%)
Frame = +3
Query: 35 AASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGAD 94
A S NR+ FP DFLFG SSAYQ EGA++ G+GPSIWDTFT HP++I S+G++ D
Sbjct: 81 AVSLNRSSFPHDFLFGTASSAYQYEGAAHEGGKGPSIWDTFTHNHPDRIVGRSNGDVAID 260
Query: 95 FYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLI 154
YHRYK D+ +MK+IG ++YRFSISWSR+ P KG +N GV +YNN+INE+++NG
Sbjct: 261 SYHRYKEDVAMMKDIGFNAYRFSISWSRLLPRGNLKGGINQEGVIYYNNLINELISNGQT 440
Query: 155 PFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTIN 214
PF+TLFH DLPQ+LEDEY GFLSPKI +DF YA+ CF+ FGDRVKHW+TLNEP+ Y+
Sbjct: 441 PFITLFHSDLPQALEDEYGGFLSPKIEQDFADYAEVCFREFGDRVKHWITLNEPLLYSTQ 620
Query: 215 GYHGGTSPPARCSKYVGNCS 234
GY G+SPP RCSK V NC+
Sbjct: 621 GYGSGSSPPMRCSKSVANCN 680
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,763,364
Number of Sequences: 36976
Number of extensions: 293225
Number of successful extensions: 1620
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 1510
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1528
length of query: 509
length of database: 9,014,727
effective HSP length: 100
effective length of query: 409
effective length of database: 5,317,127
effective search space: 2174704943
effective search space used: 2174704943
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133864.8


