

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.6 - phase: 0
(368 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
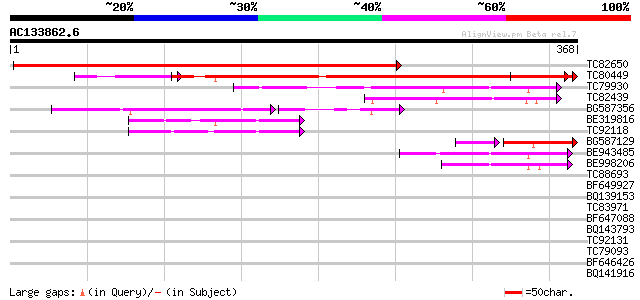
Score E
Sequences producing significant alignments: (bits) Value
TC82650 similar to GP|13374866|emb|CAC34500. putative protein {A... 509 e-145
TC80449 weakly similar to GP|15293267|gb|AAK93744.1 unknown prot... 256 2e-77
TC79930 weakly similar to PIR|T04241|T04241 hypothetical protein... 115 3e-26
TC82439 similar to GP|17065050|gb|AAL32679.1 putative protein {A... 83 1e-16
BG587356 weakly similar to GP|22535532|db hypothetical protein~s... 52 4e-14
BE319816 similar to PIR|G96707|G96 hypothetical protein T2E12.6 ... 74 1e-13
TC92118 similar to PIR|T48532|T48532 hypothetical protein T22P22... 67 8e-12
BG587129 similar to PIR|T05399|T053 hypothetical protein F10M6.7... 59 2e-11
BE943485 similar to PIR|T48532|T485 hypothetical protein T22P22.... 59 4e-09
BE998206 weakly similar to GP|20804766|dbj contains ESTs AU09611... 57 1e-08
TC88693 similar to GP|17065050|gb|AAL32679.1 putative protein {A... 34 0.079
BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis... 34 0.10
BQ139153 similar to GP|19310589|gb unknown protein {Arabidopsis ... 33 0.23
TC83971 weakly similar to GP|23315108|gb|AAN20289.1 Sequence 273... 33 0.23
BF647088 weakly similar to PIR|D86338|D86 protein F5M15.18 [impo... 33 0.23
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 32 0.30
TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity... 31 0.67
TC79093 similar to GP|4102190|gb|AAD01431.1| 35 kDa seed maturat... 31 0.67
BF646426 similar to GP|10177240|dbj emb|CAB89309.1~gene_id:MRN17... 31 0.87
BQ141916 similar to GP|19172379|gb| mannitol transporter {Apium ... 30 1.1
>TC82650 similar to GP|13374866|emb|CAC34500. putative protein {Arabidopsis
thaliana}, partial (41%)
Length = 784
Score = 509 bits (1311), Expect = e-145
Identities = 251/252 (99%), Positives = 251/252 (99%)
Frame = +3
Query: 3 SNPYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSS 62
SNPYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSS
Sbjct: 3 SNPYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSS 182
Query: 63 HPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLL 122
HPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLL
Sbjct: 183 HPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLL 362
Query: 123 RSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFD 182
RSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFD
Sbjct: 363 RSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFD 542
Query: 183 YIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMMPEVPFE 242
YIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMMPE PFE
Sbjct: 543 YIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMMPESPFE 722
Query: 243 KFRVGSQFFTLT 254
KFRVGSQFFTLT
Sbjct: 723 KFRVGSQFFTLT 758
>TC80449 weakly similar to GP|15293267|gb|AAK93744.1 unknown protein
{Arabidopsis thaliana}, partial (56%)
Length = 1239
Score = 256 bits (654), Expect(2) = 2e-77
Identities = 135/260 (51%), Positives = 170/260 (64%), Gaps = 2/260 (0%)
Frame = +3
Query: 106 LFNVYVHSDPRVNLTLLRSSNNYNPIF--KFISSKKTYRASPTLISATRRLLASAILDDA 163
LFN+Y+H+DP ++ + +F +FISSK T RASP+LISA RRLLASA+LDD
Sbjct: 165 LFNIYIHADPTTSVV------SPGGVFHNRFISSKPTQRASPSLISAARRLLASALLDDP 326
Query: 164 SNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRL 223
N YF ++SQ+C+PL SF ++Y LF + L L Y SFIEI++ P L
Sbjct: 327 LNQYFALVSQHCVPLLSFRFVYNYLFKNQLMSLASFSD----FNLLYPSFIEILSEDPNL 494
Query: 224 WKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYP 283
++RY ARG M+PEVPFE FRVGSQFF L RKHA VVV+D LW+KF++PC D CYP
Sbjct: 495 YERYNARGENVMLPEVPFEDFRVGSQFFILNRKHAKVVVRDYKLWKKFRIPCVNLDSCYP 674
Query: 284 EEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESF 343
EEHYFPTLLSMED +G TG+TLT VNWTG +GHPH Y PEE P ++ +E
Sbjct: 675 EEHYFPTLLSMEDLNGCTGFTLTRVNWTGCWDGHPHLYTPEEGFP------GANSTAEGV 836
Query: 344 LFARKFVPDCLEPLMGIAKS 363
F F+ C E L G++ S
Sbjct: 837 EF-ELFISICEEILAGVSCS 893
Score = 58.9 bits (141), Expect = 3e-09
Identities = 29/43 (67%), Positives = 34/43 (78%)
Frame = +1
Query: 326 VSPELILRLRKSTNSESFLFARKFVPDCLEPLMGIAKSVIFKD 368
VSPELI +LR S +S S+LFARKF P+CL PLM IA VIF+D
Sbjct: 802 VSPELIRQLRVSNSSYSYLFARKFSPECLAPLMDIADDVIFRD 930
Score = 51.2 bits (121), Expect(2) = 2e-77
Identities = 32/71 (45%), Positives = 39/71 (54%), Gaps = 1/71 (1%)
Frame = +2
Query: 43 DDINLFNTAISHSTPPSSSSHPSKFFHLSS-KNPTFKIAFLFLTNTDLHFTPLWNLFFQT 101
DD+ LF+ A +H+T HLS+ NP KIAFLFLTNT+L F PLW FF
Sbjct: 8 DDLALFHRATTHTTT-----------HLSTLTNPKPKIAFLFLTNTNLTFAPLWEKFFTG 154
Query: 102 TPSKLFNVYVH 112
L Y+H
Sbjct: 155 NQPSL--QYIH 181
>TC79930 weakly similar to PIR|T04241|T04241 hypothetical protein F14M19.150
- Arabidopsis thaliana, partial (53%)
Length = 1154
Score = 115 bits (288), Expect = 3e-26
Identities = 73/241 (30%), Positives = 122/241 (50%), Gaps = 28/241 (11%)
Frame = +2
Query: 146 TLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFG 205
+++ A RRLLA+A+LD N +F++LS CIP+ FD++Y L L+
Sbjct: 98 SMVEAERRLLANALLDP-DNQHFVLLSDSCIPVRRFDFVYNYLLLTSV------------ 238
Query: 206 VRLKYKSFIE-IINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKD 264
SFI+ ++ GP RY R M+PEV + FR GSQ+F++ R+HAL+++ D
Sbjct: 239 ------SFIDSYVDTGPHGNGRYVER----MLPEVEKKDFRKGSQWFSMKRQHALIIMAD 388
Query: 265 RTLWRKFKVPCYRDDE----CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHT 320
+ KFK C + E CY +EHY PT +M D G+ +++T V+W+ HP +
Sbjct: 389 SLYFNKFKHHCRPNMEGNRNCYSDEHYLPTFFNMLDPGGIANWSVTYVDWS-EGKWHPRS 565
Query: 321 YQPEEVSPELILRLR-----------------------KSTNSESFLFARKFVPDCLEPL 357
++ +++ +L+ ++ + +LFARKF P+ + L
Sbjct: 566 FRARDITYKLMKKIAYIDESPHYTSDAKRTVVIQPCELNGSKRSCYLFARKFYPETQDKL 745
Query: 358 M 358
+
Sbjct: 746 I 748
>TC82439 similar to GP|17065050|gb|AAL32679.1 putative protein {Arabidopsis
thaliana}, partial (47%)
Length = 1019
Score = 83.2 bits (204), Expect = 1e-16
Identities = 48/156 (30%), Positives = 81/156 (51%), Gaps = 28/156 (17%)
Frame = +1
Query: 231 GRYA--MMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPC---YRDDECYPEE 285
GRY+ M+PEV + FR G+Q+F++ R+HA++V+ D + KF+ C C +E
Sbjct: 121 GRYSHRMLPEVEVKDFRKGAQWFSMKRQHAVIVMADYLYYSKFRAYCQPGLEGKNCIADE 300
Query: 286 HYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRL----------- 334
HY PT + D G+ +++T+V+W+ HP +Y+ +V+ EL+ +
Sbjct: 301 HYLPTFFQIVDPGGIANWSVTHVDWSER-KWHPKSYRDHDVTYELLKNITSVDVSVHVTS 477
Query: 335 --RKSTNS----------ESFLFARKFVPDCLEPLM 358
+K S +LFARKF P+ L+ L+
Sbjct: 478 DEKKEVQSWPCLWNGIQKPCYLFARKFTPETLDKLL 585
>BG587356 weakly similar to GP|22535532|db hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 At5g14550, partial
(45%)
Length = 846
Score = 52.4 bits (124), Expect(2) = 4e-14
Identities = 44/149 (29%), Positives = 67/149 (44%), Gaps = 4/149 (2%)
Frame = +3
Query: 28 PPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTF----KIAFLF 83
PP + L NL S+ + + +S+ S S N F KIAFLF
Sbjct: 81 PPRVDQAKRQAFNSLQRFNLLRRRFSYLSSFTLNSNSSSTIRRIS-NVVFDGPPKIAFLF 257
Query: 84 LTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISSKKTYRA 143
L ++ LW FFQ F++YVHS P L + +++ + +S +
Sbjct: 258 LVRQNIPLDFLWGAFFQNGDVSNFSIYVHSTPGFLLDESTTRSHFFYGRQLSNSIQVLWG 437
Query: 144 SPTLISATRRLLASAILDDASNAYFIVLS 172
++I A RLL SA L+D +N F++LS
Sbjct: 438 ESSMIQA-ERLLLSAALEDPANQRFVLLS 521
Score = 42.7 bits (99), Expect(2) = 4e-14
Identities = 27/84 (32%), Positives = 40/84 (47%), Gaps = 2/84 (2%)
Frame = +1
Query: 175 CIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRY- 233
C+PL++F Y+Y + +SP KSF++ + GRY
Sbjct: 529 CVPLYNFSYVYNYIMVSP------------------KSFVDSFLD--------AKEGRYN 630
Query: 234 -AMMPEVPFEKFRVGSQFFTLTRK 256
M P++P EK+R GSQ+ TL RK
Sbjct: 631 PKMSPKIPREKWRKGSQWVTLVRK 702
>BE319816 similar to PIR|G96707|G96 hypothetical protein T2E12.6 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 535
Score = 73.6 bits (179), Expect = 1e-13
Identities = 45/116 (38%), Positives = 68/116 (57%), Gaps = 2/116 (1%)
Frame = +3
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIF--KFI 135
K+AF+FLT + PLW FF+ +++YVHS+P N S+ +P+F + I
Sbjct: 162 KVAFMFLTRGAVFLAPLWEQFFK-GHEVYYSIYVHSNPSYN-----GSHPESPVFHGRRI 323
Query: 136 SSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
SK+ + +I A RRLL++A+L D SN F+++S+ CIPL +F IY L S
Sbjct: 324 PSKEVEWGNVNMIEAERRLLSNALL-DISNQRFVLISESCIPLFNFSTIYSYLINS 488
>TC92118 similar to PIR|T48532|T48532 hypothetical protein T22P22.120 -
Arabidopsis thaliana, partial (51%)
Length = 1044
Score = 67.4 bits (163), Expect = 8e-12
Identities = 45/114 (39%), Positives = 61/114 (53%)
Frame = +1
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
KIAF+FLT L P+W F + S+ ++VYVH P SS Y + I S
Sbjct: 625 KIAFMFLTKGPLPLAPVWERFLKGHESR-YSVYVHPTPSYQAHFPPSSVFYK---RQIPS 792
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
+ ++ A RRLLA+A+L D SN FI+LS+ CIPL+ F +Y L S
Sbjct: 793 QIAEWGRMSMCDAERRLLANALL-DISNERFILLSESCIPLYKFSLVYNYLMKS 951
>BG587129 similar to PIR|T05399|T053 hypothetical protein F10M6.70 -
Arabidopsis thaliana, partial (25%)
Length = 682
Score = 58.5 bits (140), Expect(2) = 2e-11
Identities = 32/65 (49%), Positives = 40/65 (61%), Gaps = 17/65 (26%)
Frame = -2
Query: 321 YQPEEVSPELILRLRKST-----------------NSESFLFARKFVPDCLEPLMGIAKS 363
Y+ EEV PELI+RLRKS S+ FLFARKF P+ LEPL+G+A+S
Sbjct: 585 YEAEEVVPELIVRLRKSRPRYGEDGINGSDWSVSKRSDPFLFARKFSPEALEPLLGMARS 406
Query: 364 VIFKD 368
V+F D
Sbjct: 405 VLFND 391
Score = 27.3 bits (59), Expect(2) = 2e-11
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = -3
Query: 290 TLLSMEDSDGVTGYTLTNVNWTGTVNGHP 318
TL+++ D G TLT+V+WT G P
Sbjct: 680 TLVNLRDPRGCVPATLTHVDWTVNDGGSP 594
>BE943485 similar to PIR|T48532|T485 hypothetical protein T22P22.120 -
Arabidopsis thaliana, partial (20%)
Length = 487
Score = 58.5 bits (140), Expect = 4e-09
Identities = 36/120 (30%), Positives = 62/120 (51%), Gaps = 8/120 (6%)
Frame = +1
Query: 254 TRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGT 313
+RK A+ +V + ++ C R CY +EHYFPT+L+++ + ++T V+W+
Sbjct: 1 SRKLAINIVVYTAFYP*WEQYCRR--ACYVDEHYFPTMLTIQAGSALANRSVTWVDWS-R 171
Query: 314 VNGHPHTYQPEEVSPELILRLR--------KSTNSESFLFARKFVPDCLEPLMGIAKSVI 365
HP T+ +++ E R+R ++ LFARKF P LEPL+ + S +
Sbjct: 172 GGAHPATFGRTDITNEFFDRVRGGQKCLYNNRNSTLCSLFARKFAPSALEPLLDMVNSKV 351
>BE998206 weakly similar to GP|20804766|dbj contains ESTs AU096118(S12194)
AU096119(S12194)~unknown protein, partial (13%)
Length = 446
Score = 56.6 bits (135), Expect = 1e-08
Identities = 35/93 (37%), Positives = 52/93 (55%), Gaps = 8/93 (8%)
Frame = -2
Query: 281 CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLR----K 336
CY +EHY PTL+S++ +G + +L V+W+ HP + +V+ + + RLR K
Sbjct: 439 CYADEHYLPTLVSIKFGEGNSNRSLPWVDWS-RGGPHPAHFLRSDVNVKFLERLRGKKCK 263
Query: 337 STNSES----FLFARKFVPDCLEPLMGIAKSVI 365
N E FLFARKF+P L L+ IA V+
Sbjct: 262 YNNGEGPNACFLFARKFLPSTLSKLLKIAPKVM 164
>TC88693 similar to GP|17065050|gb|AAL32679.1 putative protein {Arabidopsis
thaliana}, partial (26%)
Length = 952
Score = 34.3 bits (77), Expect = 0.079
Identities = 18/36 (50%), Positives = 23/36 (63%)
Frame = +1
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHS 113
KIAF+FL+ L LW+ FFQ K F+VYVH+
Sbjct: 766 KIAFMFLSPGSLPLEKLWDNFFQGHEGK-FSVYVHA 870
>BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 633
Score = 33.9 bits (76), Expect = 0.10
Identities = 30/107 (28%), Positives = 43/107 (40%)
Frame = +2
Query: 5 PYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHP 64
P L L + +L L ++ HP + SP L+ L +S
Sbjct: 359 PLRRVLCLTLKVVVLVVLVVRLISHHPYRLGNSPIKRLNLGLLLTKFLST---------- 508
Query: 65 SKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYV 111
+ K + K+ F+FLT L F LW+ FFQ K F+VYV
Sbjct: 509 -----MLFKARSLKLLFMFLTPGTLPFEKLWHTFFQGHEGK-FSVYV 631
>BQ139153 similar to GP|19310589|gb unknown protein {Arabidopsis thaliana},
partial (31%)
Length = 673
Score = 32.7 bits (73), Expect = 0.23
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Frame = +3
Query: 19 LFFLAP-AILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHP 64
L+ L P A+L + P+P P+ E + + T ISHS+PPS +S P
Sbjct: 114 LYALLPIALLSLYLYPLPFLPSTESELPHSTTTIISHSSPPSFTSTP 254
>TC83971 weakly similar to GP|23315108|gb|AAN20289.1 Sequence 273 from
patent US 6410718, partial (41%)
Length = 683
Score = 32.7 bits (73), Expect = 0.23
Identities = 15/38 (39%), Positives = 22/38 (57%)
Frame = +2
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDP 115
K+AFLFL + PLW FF+ +++ VHS+P
Sbjct: 572 KVAFLFLVRGPVPLAPLWEKFFKGHKG-YYSIXVHSNP 682
>BF647088 weakly similar to PIR|D86338|D86 protein F5M15.18 [imported] -
Arabidopsis thaliana, partial (2%)
Length = 678
Score = 32.7 bits (73), Expect = 0.23
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 13/111 (11%)
Frame = +3
Query: 26 ILPPHPSPIPIS-----PTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTF--- 77
I PP +P P + PT L + S + P S+++ PS+ LS +P
Sbjct: 96 ISPPQHTPXPSTTLTSTPTQTLSSXTQQHLTNSLTPPSSNATKPSRKTSLSXDSPNTTPH 275
Query: 78 ----KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVN-LTLLR 123
+ F F ++T H +PL ++F Q T S L ++ S +N L+ LR
Sbjct: 276 SFFPQTXFRFQSSTS-HCSPLASVFHQQTQSPLPQKFLISLTSLNRLSXLR 425
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 32.3 bits (72), Expect = 0.30
Identities = 26/82 (31%), Positives = 33/82 (39%), Gaps = 8/82 (9%)
Frame = +3
Query: 10 FSLLLSLPILFFLAPAI----LPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPS 65
F L +P +FFL ++ +PP PSP P P + PP SSSHP
Sbjct: 126 FPLPYLIPPIFFLFLSL*RSFIPPPPSPFPPPPFSSPPPL-----------PPISSSHPP 272
Query: 66 K----FFHLSSKNPTFKIAFLF 83
+F L PT F F
Sbjct: 273 SLLLYYFILPLSPPTLSFFFPF 338
>TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity to human
PCF11p homolog (GB:AF046935) {Arabidopsis thaliana},
partial (24%)
Length = 1003
Score = 31.2 bits (69), Expect = 0.67
Identities = 15/48 (31%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Frame = +3
Query: 23 APAILPPHPSPIPISPTDE--LDDINLFNTAISHSTPPSSSSHPSKFF 68
AP+ +PPHP+P P P + + NL N+ ++ ++ PS+ F
Sbjct: 288 APSQMPPHPNPSPFVPNQQPTVGYSNLINSLMAQGVISLTNQAPSQDF 431
>TC79093 similar to GP|4102190|gb|AAD01431.1| 35 kDa seed maturation protein
{Glycine max}, partial (57%)
Length = 1127
Score = 31.2 bits (69), Expect = 0.67
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 6/42 (14%)
Frame = -1
Query: 55 STPPSS------SSHPSKFFHLSSKNPTFKIAFLFLTNTDLH 90
STPPSS SHP FF + P ++F+FL + + H
Sbjct: 1010 STPPSSFHA*IIHSHPRIFFRSTQHFPCLLLSFIFLASQETH 885
>BF646426 similar to GP|10177240|dbj emb|CAB89309.1~gene_id:MRN17.16~similar
to unknown protein {Arabidopsis thaliana}, partial (14%)
Length = 651
Score = 30.8 bits (68), Expect = 0.87
Identities = 16/48 (33%), Positives = 24/48 (49%)
Frame = +1
Query: 42 LDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDL 89
L + L N A+SH+ S F HLS + PT+ I+ + NT +
Sbjct: 271 LQTLALQNLAVSHTVLHRSIHSLKLFLHLSFQTPTWPISIIIKINTSI 414
>BQ141916 similar to GP|19172379|gb| mannitol transporter {Apium graveolens
var. dulce}, partial (9%)
Length = 824
Score = 30.4 bits (67), Expect = 1.1
Identities = 27/82 (32%), Positives = 37/82 (44%), Gaps = 9/82 (10%)
Frame = +1
Query: 5 PYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSS--- 61
PY+ ++SLL L+PAI P HP PIPI P + + IS PPS
Sbjct: 361 PYMDSYSLLP-------LSPAIPP*HPPPIPI-PIRYIYHLPSQTHPISPIIPPSLESPY 516
Query: 62 ------SHPSKFFHLSSKNPTF 77
SH + +L S + T+
Sbjct: 517 SPT*R*SHSTPIINLPSPSRTY 582
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,926,978
Number of Sequences: 36976
Number of extensions: 245006
Number of successful extensions: 2091
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 2031
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2073
length of query: 368
length of database: 9,014,727
effective HSP length: 97
effective length of query: 271
effective length of database: 5,428,055
effective search space: 1471002905
effective search space used: 1471002905
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC133862.6


