

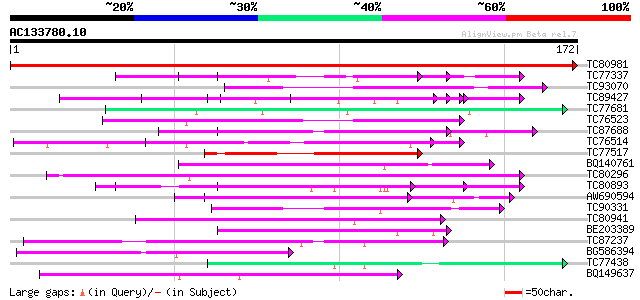
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.10 - phase: 0
(172 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|... 356 2e-99
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 58 2e-09
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 53 7e-08
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 52 1e-07
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 51 3e-07
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 50 4e-07
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 49 8e-07
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 47 3e-06
TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Ar... 47 3e-06
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 47 4e-06
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 46 6e-06
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 46 8e-06
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 45 1e-05
TC90331 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 45 2e-05
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 43 5e-05
BE203389 weakly similar to PIR|T06754|T0 DNA-directed RNA polyme... 43 5e-05
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 43 5e-05
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 43 7e-05
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 42 9e-05
BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g1261... 40 4e-04
>TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|AI994480
gb|T44186 gb|Z30806 come from this gene. {Arabidopsis
thaliana}, partial (37%)
Length = 699
Score = 356 bits (914), Expect = 2e-99
Identities = 172/172 (100%), Positives = 172/172 (100%)
Frame = +3
Query: 1 MAEPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPN 60
MAEPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPN
Sbjct: 51 MAEPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPN 230
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS
Sbjct: 231 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 410
Query: 121 DDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVRISGGTPNAD 172
DDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVRISGGTPNAD
Sbjct: 411 DDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVRISGGTPNAD 566
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 57.8 bits (138), Expect = 2e-09
Identities = 37/120 (30%), Positives = 51/120 (41%), Gaps = 18/120 (15%)
Frame = +3
Query: 33 PKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDE-----ENSDEEPVVD 87
P+L + D T + S D + E E+ DD+DE+D+D E DE D
Sbjct: 327 PELYALMLTDMPTTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSED 506
Query: 88 RKGKGIMRDDKGKGKMI-------------EEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
G G ++K K EE+DED DD D+ DD D+D D G + D
Sbjct: 507 GGGYGNNSNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEED 686
Score = 42.0 bits (97), Expect = 1e-04
Identities = 20/62 (32%), Positives = 37/62 (59%)
Frame = +3
Query: 64 ENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDD 123
EN ++DDED +D++ D+E D DD+ +G EE++E+ D +D+++ ++D
Sbjct: 585 ENGEEEDDEDGDDQDEDDDEDEDD--------DDEEEGG--EEDEEEGVDEEDNEEEEED 734
Query: 124 SD 125
D
Sbjct: 735 ED 740
Score = 40.4 bits (93), Expect = 4e-04
Identities = 24/105 (22%), Positives = 50/105 (46%)
Frame = +3
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD 111
NS++ + PE DE+ E+E++ D DD+ E++DED+
Sbjct: 525 NSNNKSNSKKAPEGGAGGADENGEEEDDED-------------GDDQD-----EDDDEDE 650
Query: 112 DDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRR 156
DD D+ + ++D + V D++ ++ + D + + P + R++
Sbjct: 651 DDDDEEEGGEEDEEEGVDEEDNEEEEE---DEDEEALQPPKKRKK 776
Score = 26.2 bits (56), Expect = 6.9
Identities = 11/42 (26%), Positives = 22/42 (52%)
Frame = +3
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEE 83
DQ+ + + + E E D+++++E+EE+ DEE
Sbjct: 621 DQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEE 746
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 52.8 bits (125), Expect = 7e-08
Identities = 31/98 (31%), Positives = 46/98 (46%)
Frame = +1
Query: 66 NTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSD 125
++DD+ E+ + E++ DE+ E+EDEDDDD DD DD DDD D
Sbjct: 220 DSDDEIEEMDFEDDEDED---------------------EDEDEDDDDDDDDDDEDDDDD 336
Query: 126 GNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVR 163
G +D + D L +P R + + + GVR
Sbjct: 337 GFAEPTDLNAKDKRLK----SKTVPDRQQEKEKEKGVR 438
Score = 34.3 bits (77), Expect = 0.026
Identities = 21/93 (22%), Positives = 40/93 (42%), Gaps = 10/93 (10%)
Frame = +1
Query: 30 NKNPKLTST----------TTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEEN 79
+ NPK+ T + D + E+ + +E E++ DDDD+DDED+++
Sbjct: 154 HSNPKILKTNRKSTYGKFLSPYDSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDD 333
Query: 80 SDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDD 112
D K D + K K + + ++ +
Sbjct: 334 DGFAEPTDLNAK----DKRLKSKTVPDRQQEKE 420
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 52.0 bits (123), Expect = 1e-07
Identities = 27/79 (34%), Positives = 43/79 (54%)
Frame = +1
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
N+ +++ D+D DED+E++D E G G DD E++D+DDDD+DD +D
Sbjct: 346 NDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDD------EDDDDDDDDNDDGEDE 507
Query: 121 DDDSDGNVSGSDSDFSDDP 139
D+D + + D D P
Sbjct: 508 DEDEE------EDDDEDQP 546
Score = 50.1 bits (118), Expect = 4e-07
Identities = 30/104 (28%), Positives = 50/104 (47%), Gaps = 8/104 (7%)
Frame = +1
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD---- 116
N+ + T+DDD++D+D++ +DE+ DD + +E+DED D DD
Sbjct: 289 NKDGSETEDDDDEDDDDDVNDED------------DDNDEDFSGDEDDEDADPEDDPVPN 432
Query: 117 ----SDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRR 156
SDD D+D D + +D +D E D D P +++
Sbjct: 433 GAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSKKKK 564
Score = 49.7 bits (117), Expect = 6e-07
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Frame = +1
Query: 41 VDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDK-- 98
+DQ ++ T + D + ++ DDDD +DED++N ++ + DD
Sbjct: 250 LDQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVP 429
Query: 99 --GKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
G ++ED+DDDD DD+DD +D+ + D D
Sbjct: 430 NGAGGSDDDDEDDDDDD-DDNDDGEDEDEDEEEDDDED 540
Score = 46.6 bits (109), Expect = 5e-06
Identities = 21/54 (38%), Positives = 32/54 (58%)
Frame = +1
Query: 86 VDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDP 139
+D+ KG ++ K E+D+D+DD DD +D DDD+D + SG + D DP
Sbjct: 250 LDQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADP 411
Score = 43.1 bits (100), Expect = 5e-05
Identities = 27/115 (23%), Positives = 51/115 (43%)
Frame = +1
Query: 16 TKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDE 75
T+ D+++ + +++ + D++ E + D VPN ++ DD+D+DD+
Sbjct: 307 TEDDDDEDDDDDVNDEDDDNDEDFSGDEDDE---DADPEDDPVPNGAGGSDDDDEDDDDD 477
Query: 76 DEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSG 130
D++N D E E+EDE++DD +D S N+ G
Sbjct: 478 DDDNDDGED--------------------EDEDEEEDDDEDQPPSKKKK*VNIVG 582
Score = 42.4 bits (98), Expect = 9e-05
Identities = 28/91 (30%), Positives = 42/91 (45%), Gaps = 17/91 (18%)
Frame = +1
Query: 65 NNTDDDDED-DEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDE-------------- 109
N+T ++++D E E++ DE+ D + D+ G +E+DE
Sbjct: 271 NHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSG---DEDDEDADPEDDPVPNGAG 441
Query: 110 --DDDDSDDSDDSDDDSDGNVSGSDSDFSDD 138
DDDD DD DD DD+ DG D + DD
Sbjct: 442 GSDDDDEDDDDDDDDNDDGEDEDEDEEEDDD 534
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 50.8 bits (120), Expect = 3e-07
Identities = 42/177 (23%), Positives = 72/177 (39%), Gaps = 37/177 (20%)
Frame = +2
Query: 30 NKNPKLTSTTTVDQETELPTTVNSSS----------DAVPNNEPENNTDDDDEDDE---D 76
+K P+++ +++E EL ++S + ++E E TD +DED
Sbjct: 257 DKIPQISLDIVLEKEFELSHNSKAASVHFCGYKAYYEDNDDSEEEGETDSEDEDIPLITT 436
Query: 77 EENSDEEPVVDRKGKGIMRDDKGKG-------------KMIEEEDEDDDDSDDSDDSDDD 123
E+N EP + + K K+++ +DED DD D+SDD
Sbjct: 437 EQNGKPEPKAEELKVSEPKKADAKNAAPAKNAATAKHVKVVDPKDEDSDDDDESDDEIGS 616
Query: 124 SDGNVSGSDSDFSDD-----------PLAEVDLDNILPSRTRRRTAQSGVRISGGTP 169
SD + +DSD D+ P+ +VD P+ + +T S + TP
Sbjct: 617 SDDEMENADSDSEDEDDSDEDDEEETPVKKVDQGKKRPNESASKTPISSKKSKNATP 787
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 50.1 bits (118), Expect = 4e-07
Identities = 30/112 (26%), Positives = 50/112 (43%), Gaps = 2/112 (1%)
Frame = +3
Query: 29 PNKNPKLTSTTTVDQETELPTTVN--SSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVV 86
P + P +S D+E++ S AV ++++D DDEDD+ + D++PV
Sbjct: 144 PQRKPASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVA 323
Query: 87 DRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDD 138
+K E + + D SDD D + D DG+ S + S+D
Sbjct: 324 AKK---------------EVSESESDSSDDEDKMNVDKDGSDSDESEEESED 434
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 49.3 bits (116), Expect = 8e-07
Identities = 26/89 (29%), Positives = 45/89 (50%)
Frame = +2
Query: 46 ELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIE 105
++P V + E E DD +++ D+++ D E V +K KG +G+ E
Sbjct: 320 KVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKG---GSEGEDDFEE 490
Query: 106 EEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
E+DED++ S+D DD +D+ + G D
Sbjct: 491 EDDEDEEGSEDEDDEEDEKEKVKGGGIED 577
Score = 43.9 bits (102), Expect = 3e-05
Identities = 30/107 (28%), Positives = 53/107 (49%), Gaps = 10/107 (9%)
Frame = +2
Query: 64 ENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDD 123
E+N + +E DE +E+S+E+ +++ G+ E D++DDD DD +D + +
Sbjct: 623 EDNYEKGEERDEADEDSEEDDELEKAGEF-------------EMDDEDDDDDDEEDEEAE 763
Query: 124 SDGNVSGSD--------SDFSDDPLAEV--DLDNILPSRTRRRTAQS 160
GNV D D L EV D D++ ++ ++RTA +
Sbjct: 764 DMGNVRYEDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQKKRTAST 904
Score = 32.0 bits (71), Expect = 0.13
Identities = 19/66 (28%), Positives = 32/66 (47%), Gaps = 5/66 (7%)
Frame = +2
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPV-----VDRKGKGIMRDDKGKGKMIEE 106
+S D E DD+D+DD+DEE+ + E + D G + K K +++E
Sbjct: 668 DSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMGNVRYEDFFGGKNEKGSKRKDQLLEV 847
Query: 107 EDEDDD 112
++DD
Sbjct: 848 SGDEDD 865
Score = 29.3 bits (64), Expect = 0.82
Identities = 28/135 (20%), Positives = 52/135 (37%), Gaps = 37/135 (27%)
Frame = +2
Query: 41 VDQETELPTTVNSSSDAVPNNEPENNTDDDD-------------EDDEDEENSDEEPVVD 87
V ++ E V + + + E + DDDD E ++D E D+E
Sbjct: 335 VGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEG 514
Query: 88 RKGKGIMRDDKGKGK----------------MIEEEDEDDDDSDDSDDSDDDSD------ 125
+ + D+K K K +E+E+++ + ++ D++D+DS+
Sbjct: 515 SEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEERDEADEDSEEDDELE 694
Query: 126 --GNVSGSDSDFSDD 138
G D D DD
Sbjct: 695 KAGEFEMDDEDDDDD 739
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 47.4 bits (111), Expect = 3e-06
Identities = 30/103 (29%), Positives = 51/103 (49%), Gaps = 6/103 (5%)
Frame = +3
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKG 101
D+E + P S + + P + +++D++ D+E +E+ D +PV K K K
Sbjct: 771 DEEDKKPAAKASKNVSAPTKKAASSSDEES-DEESDEDEDAKPV----SKPAAVAKKSKK 935
Query: 102 KMIEEEDEDDDDSDDSDDS------DDDSDGNVSGSDSDFSDD 138
+ +DEDDD S D D +D + + GSDSD S++
Sbjct: 936 DSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVDKDGSDSDQSEE 1064
Score = 43.1 bits (100), Expect = 5e-05
Identities = 32/133 (24%), Positives = 62/133 (46%), Gaps = 5/133 (3%)
Frame = +3
Query: 2 AEPETNNHE-EPTFPTKRKPDQEETHHL--PNKNPKLTSTTTVDQETELPTTVN--SSSD 56
A+ E+++ + E + +KP + + ++ P K +S D+E++ S
Sbjct: 732 ADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPA 911
Query: 57 AVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD 116
AV ++++D DDEDD+ + D++PV +K E++ D D D
Sbjct: 912 AVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKK---------------EDKMNVDKDGSD 1046
Query: 117 SDDSDDDSDGNVS 129
SD S+++S+ S
Sbjct: 1047SDQSEEESEDEPS 1085
Score = 37.7 bits (86), Expect = 0.002
Identities = 40/167 (23%), Positives = 65/167 (37%), Gaps = 22/167 (13%)
Frame = +3
Query: 2 AEPETNNHEEPTFPTKRKPDQEETHH-----LPNKN-----PKLTSTTTVDQETELPTTV 51
A+PET + E + D+EE +P+KN K+ ++ D E
Sbjct: 393 AQPETTSEESDS--DSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDK 566
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRK--GKGIMRDDKGKGKMIEEEDE 109
++ AVP+ D+ED + S +E D K K + + + +
Sbjct: 567 KPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTES 746
Query: 110 DDDDSDDSDDSDD----DSDGNVS------GSDSDFSDDPLAEVDLD 146
D+DS+ SD+ D + NVS S SD D ++ D D
Sbjct: 747 SDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDED 887
Score = 37.4 bits (85), Expect = 0.003
Identities = 30/131 (22%), Positives = 54/131 (40%), Gaps = 10/131 (7%)
Frame = +3
Query: 5 ETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSS-------SDA 57
E ++ E P + P + N K T +E++ ++ + S A
Sbjct: 300 EESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKPVSKA 479
Query: 58 VPNNE---PENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDS 114
VP+ P D +E+D EE+SDE D+K K ++ D++D+
Sbjct: 480 VPSKNGSAPAKKVDTSEEEDS-EESSDE----DKKPAAKAVPSKNGSAPAKKAASDEEDT 644
Query: 115 DDSDDSDDDSD 125
D+S D D++ +
Sbjct: 645 DESSDEDEEDE 677
Score = 28.9 bits (63), Expect = 1.1
Identities = 33/174 (18%), Positives = 63/174 (35%), Gaps = 39/174 (22%)
Frame = +3
Query: 17 KRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDE- 75
K + EE+ ++ P + + + P + A P E + D DE
Sbjct: 282 KEESSSEESSESEDEQPVVKAPAP---SKKTPAKKGNVKKAQPETTSEESDSDSSSSDEE 452
Query: 76 -------------------------DEENSDEEPVVDRK--GKGI-MRDDKGKGKMIEEE 107
+EE+S+E D+K K + ++ K +
Sbjct: 453 EVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASD 632
Query: 108 DEDDDDSDDSDDSDD--------DSDGNVSG--SDSDFSDDPLAEVDLDNILPS 151
+ED D+S D D+ D+ +G+V +D++ SD+ D ++ P+
Sbjct: 633 EEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPA 794
>TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Arabidopsis
thaliana, partial (70%)
Length = 907
Score = 47.4 bits (111), Expect = 3e-06
Identities = 22/66 (33%), Positives = 40/66 (60%)
Frame = +3
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDD 119
NN+P ++ DDDE++ D++NS+ + +D+ G+ ++DE++DD D+ DD
Sbjct: 480 NNKP--SSADDDEEESDKDNSE-----------VNNEDEASGEDDHQDDEEEDDEDEDDD 620
Query: 120 SDDDSD 125
DD+ D
Sbjct: 621 EDDE*D 638
Score = 33.9 bits (76), Expect = 0.033
Identities = 17/66 (25%), Positives = 40/66 (59%), Gaps = 1/66 (1%)
Frame = +3
Query: 17 KRKPDQEETHHLPNKNPKLTSTTTVDQETELPTT-VNSSSDAVPNNEPENNTDDDDEDDE 75
KRK + E+ + N P +S ++E++ + VN+ +A ++ +++ ++DDED++
Sbjct: 441 KRKVEYEKLMNAYNNKP--SSADDDEEESDKDNSEVNNEDEASGEDDHQDDEEEDDEDED 614
Query: 76 DEENSD 81
D+E+ +
Sbjct: 615 DDEDDE 632
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 47.0 bits (110), Expect = 4e-06
Identities = 31/112 (27%), Positives = 50/112 (43%), Gaps = 16/112 (14%)
Frame = +1
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD 111
+SS + +++ +++DD + D+ +SDEEPV K + + K K + DD
Sbjct: 274 DSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVPAPKKEKAKKAKKAKSVSSSSDSSDD 453
Query: 112 D----------------DSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
+SDDS + DSD + S S SD S D + D D+
Sbjct: 454 SSSESESEPEPXNVPLPESDDSSSASSDSDSS-SDSSSDSSSDSSSXSDXDS 606
Score = 38.1 bits (87), Expect = 0.002
Identities = 22/76 (28%), Positives = 32/76 (41%)
Frame = +1
Query: 97 DKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRR 156
D + D D DSD SDDSDD SD + S SD P P + + +
Sbjct: 250 DSSDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVP---------APKKEKAK 402
Query: 157 TAQSGVRISGGTPNAD 172
A+ +S + ++D
Sbjct: 403 KAKKAKSVSSSSDSSD 450
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 46.2 bits (108), Expect = 6e-06
Identities = 34/148 (22%), Positives = 61/148 (40%), Gaps = 3/148 (2%)
Frame = +3
Query: 12 PTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNS---SSDAVPNNEPENNTD 68
PT P K KP ++ P +D + E ++ + S +A +++ N+D
Sbjct: 669 PTDP-KAKPKAYGEVNVAADVPGAELLQIIDDDVEQESSHSDDCGSDNAQEDDQVSLNSD 845
Query: 69 DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNV 128
DD++ D SD++ D G +D+ +D D+ + + D D + DG +
Sbjct: 846 DDNQLGSDNTGSDDDEAEDHDGVSDDENDRSSDYETSGDDADNVEDEGDDLEDSEEDGGI 1025
Query: 129 SGSDSDFSDDPLAEVDLDNILPSRTRRR 156
S + D L VD L ++R
Sbjct: 1026SEHEGDGDLHILGSVDTKTTLKDLAKKR 1109
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 45.8 bits (107), Expect = 8e-06
Identities = 30/99 (30%), Positives = 44/99 (44%), Gaps = 6/99 (6%)
Frame = +3
Query: 64 ENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDD------SDDS 117
EN +D +D++D+D D++ V D G D G E + EDD + SDD
Sbjct: 306 ENKSDTEDDEDDD----DDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDG 473
Query: 118 DDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRR 156
+D DDD D D D+ E D + P +R+
Sbjct: 474 EDDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPPAKKRK 590
Score = 45.4 bits (106), Expect = 1e-05
Identities = 32/116 (27%), Positives = 50/116 (42%), Gaps = 9/116 (7%)
Frame = +3
Query: 33 PKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKG-- 90
PKLT+ +D L + N + +DDD+DD+ ++ D+ D G
Sbjct: 237 PKLTA---MDSRFPLEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDE 407
Query: 91 ---KGIMRDD---KGKGKMIEEEDEDDD-DSDDSDDSDDDSDGNVSGSDSDFSDDP 139
+G DD G G + ED+DDD D +D +D +D+ D D + P
Sbjct: 408 GEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPP 575
Score = 41.6 bits (96), Expect = 2e-04
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 2/99 (2%)
Frame = +3
Query: 27 HLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVV 86
HL + P + ++ E + D +E ++ ++D DE EE D E
Sbjct: 276 HLLGRKPAYKENKSDTEDDE-----DDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDP 440
Query: 87 DRKGKGIMRDDKGKGKMIEEEDEDD--DDSDDSDDSDDD 123
+ G G D + +EEDE+D D+ D+ D+ +DD
Sbjct: 441 EANGAGGSDDGEDDDDDGDEEDEEDGEDEEDEEDEEEDD 557
Score = 27.7 bits (60), Expect = 2.4
Identities = 9/38 (23%), Positives = 21/38 (54%)
Frame = +3
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRK 89
+ D +E + ++D+ED+E+++ + + P RK
Sbjct: 477 DDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPPAKKRK 590
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 45.1 bits (105), Expect = 1e-05
Identities = 21/72 (29%), Positives = 39/72 (54%)
Frame = +3
Query: 51 VNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDED 110
V ++ E E D+ DE++EDE + +EE + + + D++ + + EEE+E+
Sbjct: 42 VEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEE 221
Query: 111 DDDSDDSDDSDD 122
DDD + +D D
Sbjct: 222 DDDEEGEEDEID 257
Score = 43.1 bits (100), Expect = 5e-05
Identities = 24/95 (25%), Positives = 50/95 (52%), Gaps = 1/95 (1%)
Frame = +3
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDD 119
N E + +++ED+ DEE DE + + + +D+ ++E+E++++ ++ ++
Sbjct: 54 NEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEH-----DDEEEEEEEEEEEEE 218
Query: 120 SDDDSDGNVSGSDS-DFSDDPLAEVDLDNILPSRT 153
DDD +G D DPL + + + LP+RT
Sbjct: 219 EDDDEEGEEDEIDRISTLPDPLL-LHILSFLPTRT 320
Score = 37.0 bits (84), Expect = 0.004
Identities = 19/89 (21%), Positives = 42/89 (46%)
Frame = +3
Query: 62 EPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSD 121
E N +D+ E +E+E+ DEE ++ + EEE+E++++ + D+ +
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEE------------EEDEHDEEEEEEEEEEEEDEHDDEEE 188
Query: 122 DDSDGNVSGSDSDFSDDPLAEVDLDNILP 150
++ + + D + E+D + LP
Sbjct: 189 EEEEEEEEEEEDDDEEGEEDEIDRISTLP 275
Score = 28.9 bits (63), Expect = 1.1
Identities = 21/88 (23%), Positives = 36/88 (40%), Gaps = 4/88 (4%)
Frame = +3
Query: 5 ETNNHEEPTFPTKRKPDQ----EETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPN 60
E +N EE + + D+ EE H D+E E +
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEH--------------DEEEEEEEEEEEEDEHDDE 182
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDR 88
E E ++++E+D+DEE ++E +DR
Sbjct: 183 EEEEEEEEEEEEEDDDEEGEEDE--IDR 260
>TC90331 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (25%)
Length = 557
Score = 44.7 bits (104), Expect = 2e-05
Identities = 27/92 (29%), Positives = 46/92 (49%), Gaps = 3/92 (3%)
Frame = -1
Query: 62 EPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD---DDSDDSD 118
E E++ DD D+DD + E+ DEE +D G G +++ +DD D + D D
Sbjct: 554 EEEDDDDDGDDDDGEFEDDDEE------------EDFGAGYLVQPVGQDDPLNDGAADID 411
Query: 119 DSDDDSDGNVSGSDSDFSDDPLAEVDLDNILP 150
D +++ DG + D D+ + + D +LP
Sbjct: 410 DGEENEDGEEEEEEVD--DEDVDDDDAQEVLP 321
Score = 32.7 bits (73), Expect = 0.074
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Frame = -1
Query: 104 IEEEDEDDDDSDDSDDSDDDSDGNVSGS----DSDFSDDPLAE--VDLDN 147
+EEED+DDD DD + +DD + G+ DDPL + D+D+
Sbjct: 557 VEEEDDDDDGDDDDGEFEDDDEEEDFGAGYLVQPVGQDDPLNDGAADIDD 408
Score = 26.9 bits (58), Expect = 4.1
Identities = 18/63 (28%), Positives = 27/63 (42%)
Frame = -1
Query: 51 VNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDED 110
++ + E E DD+D DD+D + K K RD+ +E DED
Sbjct: 416 IDDGEENEDGEEEEEEVDDEDVDDDDAQEVLPPASSHPKRK---RDND------DEADED 264
Query: 111 DDD 113
D+D
Sbjct: 263 DED 255
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 43.1 bits (100), Expect = 5e-05
Identities = 25/95 (26%), Positives = 45/95 (47%), Gaps = 1/95 (1%)
Frame = +2
Query: 39 TTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDK 98
TT D+E + ++ + E N ++ E +E EE D E + + D+K
Sbjct: 95 TTEDKEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEGDEK 274
Query: 99 GKGKM-IEEEDEDDDDSDDSDDSDDDSDGNVSGSD 132
K + EEE+E D+D+ D +D ++G+ G +
Sbjct: 275 EKTEAETEEEEEADEDTIDKSKEEDKAEGSKGGKE 379
Score = 30.8 bits (68), Expect = 0.28
Identities = 19/71 (26%), Positives = 35/71 (48%)
Frame = +3
Query: 64 ENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDD 123
+N+ + ++ D+ E +S +EPV +DK I+E +EDD + +S++ D
Sbjct: 375 KNDNEVEEVKDDMEVDSVKEPV----------EDKNDDDDIQEMEEDDKNVAESENEKMD 524
Query: 124 SDGNVSGSDSD 134
D V + D
Sbjct: 525 VDAEVKETTED 557
>BE203389 weakly similar to PIR|T06754|T0 DNA-directed RNA polymerase I 190K
chain homolog F15B8.150 - Arabidopsis thaliana, partial
(6%)
Length = 652
Score = 43.1 bits (100), Expect = 5e-05
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Frame = +2
Query: 64 ENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD---SDDS 120
+N DDD++ D E+++E+ +K K D+ EEE D + S+D S+D
Sbjct: 401 QNGQTDDDDEVGDTEDAEEDGFDAQKSKQRATDEVDYDDGPEEETHDGEKSEDVEVSEDG 580
Query: 121 DDDSDGN---VSGSDSD 134
DD D N V+G DSD
Sbjct: 581 KDDEDDNGVEVNGDDSD 631
Score = 38.1 bits (87), Expect = 0.002
Identities = 30/102 (29%), Positives = 49/102 (47%), Gaps = 21/102 (20%)
Frame = +2
Query: 52 NSSSDAVPN---NEPENN----TDDDDE----DDEDEENSDEEPVVDRKGKGIMRDD--- 97
++SS+ + N NE +N TDDDDE +D +E+ D + R + DD
Sbjct: 347 SNSSNGLDNDHSNESASNQNGQTDDDDEVGDTEDAEEDGFDAQKSKQRATDEVDYDDGPE 526
Query: 98 -------KGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSD 132
K + + E+ +DD+D + + + DDSD V+ SD
Sbjct: 527 EETHDGEKSEDVEVSEDGKDDEDDNGVEVNGDDSDIEVNDSD 652
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 43.1 bits (100), Expect = 5e-05
Identities = 34/135 (25%), Positives = 57/135 (42%), Gaps = 6/135 (4%)
Frame = +1
Query: 5 ETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPE 64
E +H E + EE HL N+ K D+E E E
Sbjct: 55 EEKSHLENEESKETGESSEEKSHLENEENK-------DEEKSKQENEEIKDGEKIQQENE 213
Query: 65 NNTDDDDEDDEDEENSDEEPVVD----RKGKGIMRDDKGKGKMIEE--EDEDDDDSDDSD 118
N D++ E+EEN DEE +K +G +K G++ EE + E+++ S+ +
Sbjct: 214 ENKDEEKSQQENEENKDEEKSQQENELKKNEG---GEKETGEITEEKSKQENEETSETNS 384
Query: 119 DSDDDSDGNVSGSDS 133
++ + N +GSD+
Sbjct: 385 KDKENEESNQNGSDA 429
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 42.7 bits (99), Expect = 7e-05
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 3/87 (3%)
Frame = +1
Query: 3 EPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPT---TVNSSSDAVP 59
E ET E PTK + D ET KN + +TT + ETE PT T ++++ P
Sbjct: 286 EAETTTEPETNTPTKSETDVPETTTTLEKNQNVPGSTT-ESETETPTKSETETTTTEETP 462
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVV 86
+PE + E + + EP V
Sbjct: 463 TEKPETDIPKTTTASEKNKTTTTEPTV 543
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 -
garden pea, partial (93%)
Length = 1607
Score = 42.4 bits (98), Expect = 9e-05
Identities = 36/122 (29%), Positives = 48/122 (38%), Gaps = 13/122 (10%)
Frame = +2
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDD---------KGKGKMIEE----E 107
N PE DD+D D++ E + D +RD G+ EE +
Sbjct: 860 NPPEVPEDDEDIDEDMAEELQNQMEQDYDIGSTIRDKIIPHAVSWFTGEAAQGEEFGDLD 1039
Query: 108 DEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVRISGG 167
DEDDD+ DD++D D+D D D D DD E S + AQ G G
Sbjct: 1040DEDDDEDDDAEDDDEDED-----EDEDEDDDEDEEETKTKKKSSAKKSGIAQLGEGQQGE 1204
Query: 168 TP 169
P
Sbjct: 1205RP 1210
Score = 37.7 bits (86), Expect = 0.002
Identities = 21/100 (21%), Positives = 44/100 (44%)
Frame = +2
Query: 21 DQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENS 80
D++ L N+ + + ++ +P V+ + E + DD+D+D++D+
Sbjct: 896 DEDMAEELQNQMEQDYDIGSTIRDKIIPHAVSWFTGEAAQGEEFGDLDDEDDDEDDDAED 1075
Query: 81 DEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
D+E E+EDED+DD +D +++
Sbjct: 1076DDED--------------------EDEDEDEDDDEDEEET 1135
Score = 32.3 bits (72), Expect = 0.097
Identities = 13/36 (36%), Positives = 23/36 (63%)
Frame = +2
Query: 56 DAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGK 91
D ++E ++ DDD+++DEDE+ D+E + K K
Sbjct: 1037 DDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTK 1144
Score = 31.6 bits (70), Expect = 0.17
Identities = 13/49 (26%), Positives = 27/49 (54%)
Frame = +2
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGK 100
+ D +++ + + D+D++DDEDEE + + K GI + +G+
Sbjct: 1049 DDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKSGIAQLGEGQ 1195
Score = 28.5 bits (62), Expect = 1.4
Identities = 9/39 (23%), Positives = 24/39 (61%)
Frame = +2
Query: 51 VNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRK 89
++ D ++ +++ D+D+++DED++ +EE +K
Sbjct: 1034 LDDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKK 1150
>BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g12610 [imported]
- Arabidopsis thaliana, partial (15%)
Length = 538
Score = 40.4 bits (93), Expect = 4e-04
Identities = 28/127 (22%), Positives = 53/127 (41%), Gaps = 17/127 (13%)
Frame = +3
Query: 10 EEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTT-----VNSSSDAVPNNEPE 64
E+ K++P+++ T + P+ D + E + + D N+PE
Sbjct: 30 EDEEVDRKKRPNKKNTFDDDEEGPRGGDHDEDDYDVEKGDDWEHEEIFTDDDEAVGNDPE 209
Query: 65 NNTD------------DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDD 112
D DDE++E++E ++E + + GK + + G + E +D+DD
Sbjct: 210 EREDLAPEVPAPPEIKQDDEEEEEDEXNEEGGGLSKSGKELKKLLGRTGGLSESDDDDDX 389
Query: 113 DSDDSDD 119
D DD
Sbjct: 390 XDDXIDD 410
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.298 0.124 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,223,281
Number of Sequences: 36976
Number of extensions: 62936
Number of successful extensions: 3348
Number of sequences better than 10.0: 448
Number of HSP's better than 10.0 without gapping: 1011
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2001
length of query: 172
length of database: 9,014,727
effective HSP length: 89
effective length of query: 83
effective length of database: 5,723,863
effective search space: 475080629
effective search space used: 475080629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC133780.10


