

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.10 + phase: 2 /pseudo
(609 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
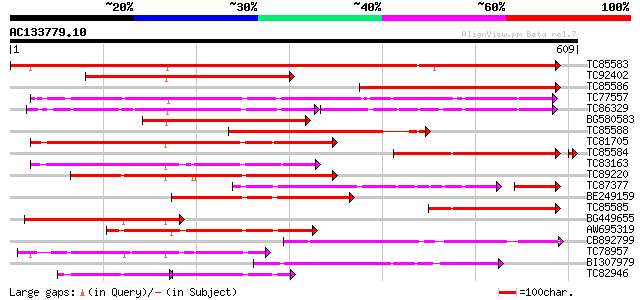
Score E
Sequences producing significant alignments: (bits) Value
TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase ... 879 0.0
TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like ser... 449 e-126
TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin... 423 e-119
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 392 e-109
TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 204 8e-94
BG580583 weakly similar to GP|9757901|dbj serine protease-like p... 307 9e-84
TC85588 similar to GP|20198169|gb|AAM15440.1 subtilisin-like ser... 275 4e-74
TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 273 2e-73
TC85584 weakly similar to PIR|A84473|A84473 probable serine prot... 268 3e-72
TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 258 4e-69
TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 249 3e-66
TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like prot... 182 3e-59
BE249159 weakly similar to GP|20198252|gb subtilisin-like serine... 218 5e-57
TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin... 205 4e-53
BG449655 similar to GP|20198252|gb subtilisin-like serine protea... 201 9e-52
AW695319 similar to GP|6708179|gb| subtilisin-type serine endope... 193 2e-49
CB892799 weakly similar to PIR|T05768|T05 subtilisin-like protei... 187 8e-48
TC78957 similar to GP|14091078|gb|AAK53589.1 subtilisin-like pro... 183 1e-46
BI307979 weakly similar to GP|20521377|db putative subtilisin-li... 177 1e-44
TC82946 weakly similar to GP|20160863|dbj|BAB89802. putative sub... 89 2e-38
>TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase AIR3
auxin-induced [imported] - Arabidopsis thaliana
(fragment), partial (40%)
Length = 2691
Score = 879 bits (2270), Expect = 0.0
Identities = 448/621 (72%), Positives = 502/621 (80%), Gaps = 31/621 (4%)
Frame = +2
Query: 2 SFH*FHKLLVFYYYFFFLFQ-------CYIVYMGAHSHGPTPTSVDLETATSSHYDLLGS 54
SF H ++ + F FL + CYIVY+GAHSHGP PTS++LE AT+SHYDLL S
Sbjct: 38 SFFSLHTMVTLLFLFMFLLETVHGTKKCYIVYLGAHSHGPRPTSLELEIATNSHYDLLSS 217
Query: 55 IVGSKEEAKEAIIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEF 114
+GS+E+AKEAIIYSYNK INGFAA+LE+EEAA +AK VVSVFLSK HKLHTTRSWEF
Sbjct: 218 TLGSREKAKEAIIYSYNKHINGFAALLEDEEAADIAKKRNVVSVFLSKPHKLHTTRSWEF 397
Query: 115 LGLHGNDINSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSS------ 168
LGL N N+AWQKG+FGENTIIANIDTGVWPES+SF+D+G GP+P+KWRGG +
Sbjct: 398 LGLRRNAKNTAWQKGKFGENTIIANIDTGVWPESKSFNDKGYGPVPSKWRGGKACEISKF 577
Query: 169 ---KKVPCNRKLIGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASI 225
KK PCNRKLIGARFFS+AYE YN KLP+ QRTARDF+GHGTHTLSTAGGNFVP AS+
Sbjct: 578 SKYKKNPCNRKLIGARFFSNAYEAYNDKLPSWQRTARDFLGHGTHTLSTAGGNFVPDASV 757
Query: 226 FNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPS 285
F IGNGT+KGGSPRARVATYKVCWSL D CFGADVL+AIDQAI DGVDIIS+S G S
Sbjct: 758 FAIGNGTVKGGSPRARVATYKVCWSLLDLEDCFGADVLAAIDQAISDGVDIISLSLAGHS 937
Query: 286 STNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFS 345
E+IFTDEVSIGAFHAL+RNILLVASAGNEGPT GSVVNVAPWVFT+AAST+DRDFS
Sbjct: 938 LVYPEDIFTDEVSIGAFHALSRNILLVASAGNEGPTGGSVVNVAPWVFTIAASTLDRDFS 1117
Query: 346 STITIGDQIIRGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKI 405
STITIG+Q IRGASLFV+LPPNQ+F L+ S D K +NAT DA+FC+P TLDPSKVKGKI
Sbjct: 1118STITIGNQTIRGASLFVNLPPNQAFPLIVSTDGKLANATNHDAQFCKPGTLDPSKVKGKI 1297
Query: 406 VACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTV----------- 454
V C REG IKSVAEGQEALSAGAKGM L NQPK G T L+EPH LS V
Sbjct: 1298VECIREGNIKSVAEGQEALSAGAKGMLLSNQPK-QGKTTLAEPHTLSCVEVPHHAPKPPK 1474
Query: 455 ---GGNGQAAITAPPRLGVTATDT-IESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQV 510
+ A + P +T+ D+ +++GT I+FS A TL GRKPAPVMASFSSRGPN++
Sbjct: 1475PKKSAEQERAGSHAPAFDITSMDSKLKAGTTIKFSGAKTLYGRKPAPVMASFSSRGPNKI 1654
Query: 511 QPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIK 570
QP ILKPDVTAPGVNILAAYSL+ASASNL TDNR FPFNV+QGTSMSCPHVAG AGLIK
Sbjct: 1655QPSILKPDVTAPGVNILAAYSLYASASNLKTDNRNNFPFNVLQGTSMSCPHVAGIAGLIK 1834
Query: 571 TLHPNWSPAAIKSAIMTTVIT 591
TLHPNWSPAAIKSAIMTT T
Sbjct: 1835TLHPNWSPAAIKSAIMTTATT 1897
>TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like serine
protease AIR3 {Arabidopsis thaliana}, partial (16%)
Length = 704
Score = 449 bits (1155), Expect = e-126
Identities = 223/234 (95%), Positives = 224/234 (95%), Gaps = 9/234 (3%)
Frame = +1
Query: 82 EEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANID 141
EEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANID
Sbjct: 1 EEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANID 180
Query: 142 TGVWPESRSFSDRGIGPIPAKWRGGS---------SKKVPCNRKLIGARFFSDAYERYNG 192
TGVWPESRSFSDRGIGPIPAKWRGG+ SKKVPCNRKLIGARFFSDAYERYNG
Sbjct: 181 TGVWPESRSFSDRGIGPIPAKWRGGNVCQINKLRGSKKVPCNRKLIGARFFSDAYERYNG 360
Query: 193 KLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLT 252
KLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLT
Sbjct: 361 KLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLT 540
Query: 253 DAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALA 306
DAASCFGADVLSAIDQAIDDGVDII VSAGGPSSTNSEEIFTDEVSIGAFHALA
Sbjct: 541 DAASCFGADVLSAIDQAIDDGVDIIFVSAGGPSSTNSEEIFTDEVSIGAFHALA 702
>TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin-like
protease {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 1460
Score = 423 bits (1088), Expect = e-119
Identities = 214/216 (99%), Positives = 214/216 (99%)
Frame = +1
Query: 376 IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN 435
IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN
Sbjct: 1 IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN 180
Query: 436 QPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKP 495
QPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKP
Sbjct: 181 QPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKP 360
Query: 496 APVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGT 555
APVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGT
Sbjct: 361 APVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGT 540
Query: 556 SMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
SMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT T
Sbjct: 541 SMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATT 648
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 392 bits (1007), Expect = e-109
Identities = 242/575 (42%), Positives = 333/575 (57%), Gaps = 9/575 (1%)
Frame = +3
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
YIV+M A S P E T + L S+ S E ++Y+Y I+GF+ L
Sbjct: 330 YIVHM-AKSEMPE----SFEHHTLWYESSLQSVSDSAE-----MMYTYENAIHGFSTRLT 479
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANIDT 142
EEA L +++V +++LHTTR+ +FLGL + + + G ++ +DT
Sbjct: 480 PEEARLLESQTGILAVLPEVKYELHTTRTPQFLGLDKSA--DMFPESSSGNEVVVGVLDT 653
Query: 143 GVWPESRSFSDRGIGPIPAKWRGGSSK-----KVPCNRKLIGARFFSDAYERYNGKLP-- 195
GVWPES+SF+D G GPIP W+G CN+KLIGARFFS E G +
Sbjct: 654 GVWPESKSFNDAGFGPIPTTWKGACESGTNFTAANCNKKLIGARFFSKGVEAMLGPIDET 833
Query: 196 TSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAA 255
T ++ RD GHGTHT STA G+ VP AS+F +GT +G + RARVA YKVCW
Sbjct: 834 TESKSPRDDDGHGTHTSSTAAGSVVPDASLFGYASGTARGMATRARVAVYKVCWK----G 1001
Query: 256 SCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASA 315
CF +D+L+AID+AI D V+++S+S GG S + F D V+IGAF A+ + IL+ SA
Sbjct: 1002GCFSSDILAAIDKAISDNVNVLSLSLGGGMS----DYFRDSVAIGAFSAMEKGILVSCSA 1169
Query: 316 GNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI-IRGASLF-VDLPPNQSFTLV 373
GN GP+ S+ NVAPW+ TV A T+DRDF +++++G+ + G SL+ + P L+
Sbjct: 1170GNAGPSAYSLSNVAPWITTVGAGTLDRDFPASVSLGNGLNYSGVSLYRGNALPESPLPLI 1349
Query: 374 NSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFL 433
+ +A +NAT + C TL P V GKIV C R G V +G +AG GM L
Sbjct: 1350YAGNA--TNAT--NGNLCMTGTLSPELVAGKIVLCDR-GMNARVQKGAVVKAAGGLGMVL 1514
Query: 434 ENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGR 493
N +G L+++ H+L + L A T+ KI F T +G
Sbjct: 1515SN-TAANGEELVADTHLLPATAVGEREGNAIKKYLFSEAKPTV----KIVFQG--TKVGV 1673
Query: 494 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQ 553
+P+PV+A+FSSRGPN + P ILKPD+ APGVNILA +S + L D RR FN++
Sbjct: 1674EPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLAVDERR-VDFNIIS 1850
Query: 554 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
GTSMSCPHV+G A LIK+ HP+WSPAA++SA+MTT
Sbjct: 1851GTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTT 1955
>TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (91%)
Length = 2671
Score = 204 bits (519), Expect(2) = 8e-94
Identities = 127/323 (39%), Positives = 177/323 (54%), Gaps = 9/323 (2%)
Frame = +2
Query: 19 LFQCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFA 78
L + YI++M + P S D H S + S + E + Y+Y +GF+
Sbjct: 170 LKRTYIIHMDKFN---MPASFD------DHLQWYDSSLKSVSDTAETM-YTYKHVAHGFS 319
Query: 79 AMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIA 138
L +EA L K P ++SV ++LHTTR+ EFLGL I G+ E I+
Sbjct: 320 TRLTTQEADLLTKQPGILSVIPDVRYELHTTRTPEFLGLE-KTITLLPSSGKQSE-VIVG 493
Query: 139 NIDTGVWPESRSFSDRGIGPIPAKWRGGSS-----KKVPCNRKLIGARFFSDAYERYNGK 193
IDTGVWPE +SF D G+GP+P W+G CN+KL+GARFF+ YE G
Sbjct: 494 VIDTGVWPELKSFDDTGLGPVPKSWKGECETGKTFNSSNCNKKLVGARFFAKGYEAAFGP 673
Query: 194 LP--TSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSL 251
+ T ++ RD GHG+HT +TA G+ V GAS+F +GT KG + +ARVA YKVCW
Sbjct: 674 IDENTESKSPRDDDGHGSHTSTTAAGSAVAGASLFGFASGTAKGMATQARVAAYKVCW-- 847
Query: 252 TDAASCFGADVLSAIDQAIDDGVDIIS--VSAGGPSSTNSEEIFTDEVSIGAFHALARNI 309
CF +D+ +AID+AID + VS G T + + D V++G F A+ I
Sbjct: 848 --LGGCFTSDIAAAIDKAIDTQARTAAYKVSRLGGGLT---DYYKDTVAMGTFAAIEHGI 1012
Query: 310 LLVASAGNEGPTPGSVVNVAPWV 332
L+ +SAGN GP+ S+ NVAPW+
Sbjct: 1013LVSSSAGNGGPSKASLANVAPWI 1081
Score = 158 bits (400), Expect(2) = 8e-94
Identities = 107/257 (41%), Positives = 141/257 (54%), Gaps = 2/257 (0%)
Frame = +3
Query: 334 TVAASTIDRDFSSTITIGD-QIIRGASLFVD-LPPNQSFTLVNSIDAKFSNATTRDARFC 391
TV A TIDRDF + IT+G+ G SL+ LPPN LV + + ++ C
Sbjct: 1086 TVGARTIDRDFPAYITLGNGNRYNGVSLYNGKLPPNSPLPLVYAANVSQDSSDN----LC 1253
Query: 392 RPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVL 451
+L PSKV GKIV C R G ++ + AG GM L N G L+++ +L
Sbjct: 1254 STDSLIPSKVSGKIVICDRGGNPRA-EKSLVVKRAGGIGMILANNQDY-GEELVADSFLL 1427
Query: 452 STVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQ 511
+A+ + A+ KI F T G +P+PV+A+FSSRGPN +
Sbjct: 1428 PAAALGEKAS----NEIKKYASSAPNPTAKIAFGG--TRFGVQPSPVVAAFSSRGPNILT 1589
Query: 512 PYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKT 571
P ILKPD+ APGVNILA +S + L D R FN++ GTSMSCPHV+G A L+K
Sbjct: 1590 PKILKPDLIAPGVNILAGWSGKVGPTGLSVDTRH-VSFNIISGTSMSCPHVSGLAALLKG 1766
Query: 572 LHPNWSPAAIKSAIMTT 588
HP WSPAAI+SA+MTT
Sbjct: 1767 AHPEWSPAAIRSALMTT 1817
>BG580583 weakly similar to GP|9757901|dbj serine protease-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 592
Score = 307 bits (786), Expect = 9e-84
Identities = 149/190 (78%), Positives = 164/190 (85%), Gaps = 9/190 (4%)
Frame = +3
Query: 143 GVWPESRSFSDRGIGPIPAKWRGGS---------SKKVPCNRKLIGARFFSDAYERYNGK 193
GVWPES SF+DRGIGPIP +WRGG+ SKKVPCNRKLIGARFF+ AYE ++GK
Sbjct: 21 GVWPESESFNDRGIGPIPLRWRGGNICQLDKLNTSKKVPCNRKLIGARFFNKAYEAFHGK 200
Query: 194 LPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTD 253
LP+SQ+TARDFVGHGTHTLSTAGGNFV A+IF IGNGTIKGGSPR+RVATYK CWSLTD
Sbjct: 201 LPSSQQTARDFVGHGTHTLSTAGGNFVQNATIFGIGNGTIKGGSPRSRVATYKACWSLTD 380
Query: 254 AASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVA 313
CFGADVL+AIDQ+I DG D+ISVSAGG +TN E IFTDE+SIGAFHALARNILLVA
Sbjct: 381 VVDCFGADVLAAIDQSIYDGADLISVSAGGKPNTNPEVIFTDEISIGAFHALARNILLVA 560
Query: 314 SAGNEGPTPG 323
SAGNEGPTPG
Sbjct: 561 SAGNEGPTPG 590
>TC85588 similar to GP|20198169|gb|AAM15440.1 subtilisin-like serine
protease AIR3 {Arabidopsis thaliana}, partial (7%)
Length = 572
Score = 275 bits (703), Expect = 4e-74
Identities = 146/217 (67%), Positives = 160/217 (73%)
Frame = +1
Query: 236 GSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTD 295
GSPRARVATYKVCWSL D CFGADVL+AIDQAI DGVDIIS+S G S E+IFTD
Sbjct: 1 GSPRARVATYKVCWSLLDLEDCFGADVLAAIDQAISDGVDIISLSLAGHSLVYPEDIFTD 180
Query: 296 EVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQII 355
EVSIGAFHAL+RNILLVASAGNEGPT GSVVNV PWVFT+AAST+DRDFSSTITIG+Q I
Sbjct: 181 EVSIGAFHALSRNILLVASAGNEGPTGGSVVNVTPWVFTIAASTLDRDFSSTITIGNQTI 360
Query: 356 RGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIK 415
RGASLFV+LPPNQ+F L+ S D K +NAT DA+FC+P TLDPSKV
Sbjct: 361 RGASLFVNLPPNQAFPLIVSTDGKLANATNHDAQFCKPGTLDPSKV-------------- 498
Query: 416 SVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLS 452
KGM L NQPK G T L+EPH LS
Sbjct: 499 -------------KGMLLSNQPK-QGKTTLAEPHTLS 567
>TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (33%)
Length = 1443
Score = 273 bits (697), Expect = 2e-73
Identities = 154/339 (45%), Positives = 205/339 (60%), Gaps = 9/339 (2%)
Frame = +1
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
Y+VYMG + +G D + S H +LL SI+ S+E + +I+ YN +GF+AML
Sbjct: 85 YVVYMGNNINGE-----DDQIPESVHIELLSSIIPSEESERIKLIHHYNHAFSGFSAMLT 249
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLG--LHGNDINSAWQKGRFGENTIIANI 140
+ EA+ L+ + VVSVF +LHTTRSW+FL L + + IIA I
Sbjct: 250 QSEASALSGHDGVVSVFPDPILELHTTRSWDFLDSDLGMKPSTNVLTHQHSSNDIIIALI 429
Query: 141 DTGVWPESRSFSDRGIGPIPAKWRG-----GSSKKVPCNRKLIGARFFS--DAYERYNGK 193
DTG+WPES SF+D GIG IP+ W+G KK CNRKLIGAR+++ D +
Sbjct: 430 DTGIWPESPSFTDEGIGKIPSVWKGICMEGHDFKKSNCNRKLIGARYYNTQDTFGSNKTH 609
Query: 194 LPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTD 253
+ ++ + RD VGHGTHT STA G V A+ + + GT +GGSP R+A YK C
Sbjct: 610 IGGAKGSPRDTVGHGTHTASTAAGVNVNNANYYGLAKGTARGGSPSTRIAAYKTC----S 777
Query: 254 AASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVA 313
C G+ +L A+D AI DGVDIIS+S G SS + D ++IGAFHA R + V
Sbjct: 778 EEGCSGSTILKAMDDAIKDGVDIISISIG-LSSLMQSDYLNDPIAIGAFHAEQRGVTAVC 954
Query: 314 SAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD 352
SAGN+GP P +VVN APW+FTVAAS IDR+F STI +G+
Sbjct: 955 SAGNDGPDPNTVVNTAPWIFTVAASNIDRNFQSTIVLGN 1071
>TC85584 weakly similar to PIR|A84473|A84473 probable serine proteinase
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1241
Score = 268 bits (685), Expect(2) = 3e-72
Identities = 139/179 (77%), Positives = 153/179 (84%)
Frame = +1
Query: 413 KIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTA 472
+IKSVAEGQEALSAGAKG+ L NQP+++G TLLSEPHVLST+ G + T L +
Sbjct: 1 EIKSVAEGQEALSAGAKGVILRNQPEINGKTLLSEPHVLSTISYPGNHSRTTGRSLDIIP 180
Query: 473 TDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSL 532
+D I+SGTK+R S A TL RKPAPVMAS+SSRGPN+VQP ILKPDVTAPGVNILAAYSL
Sbjct: 181 SD-IKSGTKLRMSPAKTLNRRKPAPVMASYSSRGPNKVQPSILKPDVTAPGVNILAAYSL 357
Query: 533 FASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
FASASNL+TD RRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT T
Sbjct: 358 FASASNLITDTRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATT 534
Score = 22.7 bits (47), Expect(2) = 3e-72
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 601 QPREITPTS 609
QPREITPTS
Sbjct: 527 QPREITPTS 553
>TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (29%)
Length = 990
Score = 258 bits (660), Expect = 4e-69
Identities = 151/317 (47%), Positives = 192/317 (59%), Gaps = 6/317 (1%)
Frame = +2
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
Y+VYMG++ +G VD + S H DLL SI+ S+E + A+I+ Y+ NGF+AML
Sbjct: 95 YVVYMGSNING-----VDGQIPESVHLDLLSSIIPSEESERIALIHHYSHAFNGFSAMLT 259
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLH-GNDINSAWQKGRFGENTIIANID 141
+ EA+ LA N VVSVF +LHTTRSW+FL G + + + II ID
Sbjct: 260 QSEASALAGNDGVVSVFEDPFLELHTTRSWDFLESDLGMRPHGILKHQHSSNDIIIGVID 439
Query: 142 TGVWPESRSFSDRGIGPIPAKWRG-----GSSKKVPCNRKLIGARFFSDAYERYNGKLPT 196
TG+WPES SF D GIG IP++W+G KK CNRKLIGAR+ YN K P
Sbjct: 440 TGIWPESPSFKDEGIGKIPSRWKGVCMEAHDFKKSNCNRKLIGARY-------YNKKDP- 595
Query: 197 SQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAAS 256
+ + RDF GHGTHT STA G V AS + + GT +GGSP AR+A YK C
Sbjct: 596 -KGSPRDFNGHGTHTASTAAGVIVNNASYYGLAKGTARGGSPSARIAAYKAC----SGEG 760
Query: 257 CFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAG 316
C G +L AID AI DGVDIIS+S G S SE + +D ++IGAFHA R +++V SAG
Sbjct: 761 CSGGTLLKAIDDAIKDGVDIISISIGFSSEFLSEYL-SDPIAIGAFHAEQRGVMVVCSAG 937
Query: 317 NEGPTPGSVVNVAPWVF 333
NEGP +VVN PW+F
Sbjct: 938 NEGPDHYTVVNTTPWIF 988
>TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (39%)
Length = 1149
Score = 249 bits (635), Expect = 3e-66
Identities = 129/300 (43%), Positives = 189/300 (63%), Gaps = 13/300 (4%)
Frame = +3
Query: 66 IIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSA 125
I+++Y+ +GF+A+L ++ A ++ +P +++VF + +LHTTRS +FLGL
Sbjct: 228 ILHTYDTAFHGFSAVLTRQQVASISNHPSILAVFEDRRRQLHTTRSPQFLGLRNQ--RGL 401
Query: 126 WQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRG---GSSKKVP--CNRKLIGA 180
W + +G + I+ DTG+WPE RSFSD +GPIP +W+G K P CNRKLIGA
Sbjct: 402 WSESDYGSDVIVGVFDTGIWPERRSFSDMNLGPIPRRWKGVCESGEKFSPRNCNRKLIGA 581
Query: 181 RFFSDAYERYNGKL----PTSQ----RTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGT 232
R+FS +E G P ++ R+ RD GHGTHT STA G + A++ +G
Sbjct: 582 RYFSKGHEVGAGSAGPLNPINETVEFRSPRDADGHGTHTASTAAGRYAFQANMSGYASGI 761
Query: 233 IKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEI 292
KG +P+AR+A YKVCW + CF +D+L+A D A++DGVD+IS+S GG S
Sbjct: 762 AKGVAPKARLAVYKVCWK---NSGCFDSDILAAFDAAVNDGVDVISISIGGGDGIASP-Y 929
Query: 293 FTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD 352
+ D ++IG++ A++R + + +SAGN+GP+ SV N+APW+ TV A TIDRDF S I I D
Sbjct: 930 YLDPIAIGSYGAVSRGVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQIIIXD 1109
>TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (54%)
Length = 2071
Score = 182 bits (461), Expect(2) = 3e-59
Identities = 122/291 (41%), Positives = 169/291 (57%), Gaps = 2/291 (0%)
Frame = +2
Query: 240 ARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSI 299
ARVA YKVCW + CF +D+ + +D+AI+DGV+I+S+S GG + + + D ++I
Sbjct: 11 ARVAAYKVCW----LSGCFTSDIAAGMDKAIEDGVNILSMSIGG----SIMDYYRDIIAI 166
Query: 300 GAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGA 358
GAF A++ IL+ +SAGN GP+ S+ NVAPW+ TV A TIDRDF S IT+G+ + GA
Sbjct: 167 GAFTAMSHGILVSSSAGNGGPSAESLSNVAPWITTVGAGTIDRDFPSYITLGNGKTYTGA 346
Query: 359 SLFVDLPPNQSFTLVNSIDAKFSNATTRDARF-CRPRTLDPSKVKGKIVACAREGKIKSV 417
SL+ P + S V N + + C P +L SKV GKIV C R G + V
Sbjct: 347 SLYNGKPSSDSLLPV----VYAGNVSESSVGYLCIPDSLTSSKVLGKIVICERGGNSR-V 511
Query: 418 AEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIE 477
+G +AG GM L N + G L+++ H+L GQ + T L T
Sbjct: 512 EKGLVVKNAGGVGMILVNN-EAYGEELIADSHLL-PAAALGQKSSTV---LKDYVFTTKN 676
Query: 478 SGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILA 528
K+ F T + +P+PV+A+FSSRGPN + P ILKPD+ APGVNILA
Sbjct: 677 PRAKLVFGG--THLQVQPSPVVAAFSSRGPNSLTPKILKPDLIAPGVNILA 823
Score = 65.5 bits (158), Expect(2) = 3e-59
Identities = 27/49 (55%), Positives = 37/49 (75%)
Frame = +1
Query: 543 NRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
++R FN++ GTSMSCPH +G A ++K +P WSPAAI+SA+MTT T
Sbjct: 865 DKRHVNFNIISGTSMSCPHASGLAAIVKGAYPEWSPAAIRSALMTTAYT 1011
>BE249159 weakly similar to GP|20198252|gb subtilisin-like serine protease
AIR3 {Arabidopsis thaliana}, partial (24%)
Length = 661
Score = 218 bits (555), Expect = 5e-57
Identities = 115/199 (57%), Positives = 142/199 (70%), Gaps = 2/199 (1%)
Frame = +1
Query: 174 NRKLIGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTI 233
+RKLIGAR F YE Y GKL S TARD +GHG+HTLSTAGGNFV G S++ GNGT
Sbjct: 82 SRKLIGARAFYKGYEAYVGKLDASFYTARDTIGHGSHTLSTAGGNFVQGVSVYGNGNGTA 261
Query: 234 KGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIF 293
KGGSP+A VA YKVCW C ADVL+ + AI DGVD++SVS G + +F
Sbjct: 262 KGGSPKAHVAAYKVCWK----GGCSDADVLAGFEAAISDGVDVLSVSLG----MKTHNLF 417
Query: 294 TDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD- 352
TD +SIG+FHA+A I++VASAGN GP G+V NVAPW+FTVAASTIDRDF+S +T+GD
Sbjct: 418 TDSISIGSFHAVANGIVVVASAGNSGPYFGTVSNVAPWLFTVAASTIDRDFASYVTLGDN 597
Query: 353 QIIRGASL-FVDLPPNQSF 370
+ +G SL DLP ++ +
Sbjct: 598 KHFKGTSLSSKDLPTHKFY 654
>TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin-like
serine protease {Oryza sativa (japonica
cultivar-group)}, partial (21%)
Length = 1353
Score = 205 bits (522), Expect = 4e-53
Identities = 105/142 (73%), Positives = 115/142 (80%)
Frame = +2
Query: 450 VLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQ 509
V+ST+ + +IT P +T D I++ IR S A L GRKPAPVMASFSSRGPN+
Sbjct: 5 VVSTINYHDPRSITTPKGSEITPED-IKTNATIRMSPANALNGRKPAPVMASFSSRGPNK 181
Query: 510 VQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLI 569
VQPYILKPDVTAPGVNILAAYSL AS SNL+TDNRRGFPFN+ QGTSMSCPHV GTAGLI
Sbjct: 182 VQPYILKPDVTAPGVNILAAYSLLASVSNLVTDNRRGFPFNIQQGTSMSCPHVVGTAGLI 361
Query: 570 KTLHPNWSPAAIKSAIMTTVIT 591
KTLHPNWSPAAIKSAIMTT T
Sbjct: 362 KTLHPNWSPAAIKSAIMTTATT 427
>BG449655 similar to GP|20198252|gb subtilisin-like serine protease AIR3
{Arabidopsis thaliana}, partial (21%)
Length = 664
Score = 201 bits (510), Expect = 9e-52
Identities = 100/180 (55%), Positives = 124/180 (68%), Gaps = 9/180 (5%)
Frame = +1
Query: 17 FFLFQCYIVYMGAHSHGPTP-TSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQIN 75
F + Y+VY+G+HSH +SVD T SHY+ LGS +GS + AKE+I YSY + IN
Sbjct: 91 FAEIKSYVVYLGSHSHDSEELSSVDFNRVTDSHYEFLGSFLGSSKTAKESIFYSYTRHIN 270
Query: 76 GFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGN----DINSAWQKGRF 131
GFAA LEEE AA++AK+PKV+SVF + KLHTT SW F+GL + +S W K RF
Sbjct: 271 GFAATLEEEVAAEIAKHPKVLSVFENNGRKLHTTHSWGFMGLEDSYGVIPSSSIWNKARF 450
Query: 132 GENTIIANIDTGVWPESRSFSDRGIGPIPAKWRG----GSSKKVPCNRKLIGARFFSDAY 187
G+ IIAN+DTGVWPES+SFSD G GPIP+KWRG G CNRKLIGAR+F+ Y
Sbjct: 451 GDGIIIANLDTGVWPESKSFSDEGFGPIPSKWRGICDKGRDPSFHCNRKLIGARYFNKGY 630
>AW695319 similar to GP|6708179|gb| subtilisin-type serine endopeptidase XSP1
{Arabidopsis thaliana}, partial (27%)
Length = 648
Score = 193 bits (490), Expect = 2e-49
Identities = 108/230 (46%), Positives = 144/230 (61%), Gaps = 4/230 (1%)
Frame = +2
Query: 105 KLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWR 164
KLHTTRSW+F+GL +A +K + +TI+A +DTG+ PE +SF D G GP PAKW+
Sbjct: 5 KLHTTRSWDFIGLP----LTAKRKLKSEGDTIVALLDTGITPEFQSFKDDGFGPPPAKWK 172
Query: 165 GGSSKKVP---CNRKLIGARFFSDAYERYNGKL-PTSQRTARDFVGHGTHTLSTAGGNFV 220
G K V CN K+IGA++F + +G+ P+ + D GHGTHT STA GN V
Sbjct: 173 GTCDKYVNFSGCNNKIIGAKYF-----KLDGRSNPSDILSPIDVEGHGTHTASTAAGNIV 337
Query: 221 PGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVS 280
P AS+F + G +G AR+A YK+CW+ C D+L+A + AI DGVD+ISVS
Sbjct: 338 PNASLFGLAKGMARGAVHSARLAIYKICWT---EDGCADMDILAAFEAAIHDGVDVISVS 508
Query: 281 AGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAP 330
GG +E D ++IGAFHA+ + I+ VASAGN GPT +VVN AP
Sbjct: 509 LGG----GNENYAQDSIAIGAFHAMRKGIITVASAGNGGPTMATVVNNAP 646
>CB892799 weakly similar to PIR|T05768|T05 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (20%)
Length = 875
Score = 187 bits (476), Expect = 8e-48
Identities = 123/307 (40%), Positives = 176/307 (57%), Gaps = 6/307 (1%)
Frame = +2
Query: 295 DEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI 354
D +SIG+FHA R + +V+SAGNEG GS N+APW+ TVAA + DRDF+S I +G+
Sbjct: 2 DAISIGSFHAANRGLFVVSSAGNEGNL-GSATNLAPWMLTVAAGSTDRDFTSDIILGNGA 178
Query: 355 -IRGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVAC--ARE 411
I G SL + N S ++++ +A T + +C +L+ +K KGK++ C
Sbjct: 179 KITGESLSL-FEMNASTRIISASEAFAGYFTPYQSSYCLESSLNKTKTKGKVLVCRHVER 355
Query: 412 GKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVL-STVGGNGQAAITAPPRLGV 470
VA+ + AG GM L ++ ++ P V+ S + G + G
Sbjct: 356 STESKVAKSKIVKEAGGVGMILIDETDQD----VAIPFVIPSAIVGK---------KKGQ 496
Query: 471 TATDTIESGTK--IRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILA 528
+++ K + +A T+IG + AP +A+FSSRGPN + P ILKPD+TAPG+NILA
Sbjct: 497 KILSYLKTTRKPMSKILRAKTVIGAQSAPRVAAFSSRGPNALNPEILKPDITAPGLNILA 676
Query: 529 AYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
A+S A G FN++ GTSM+CPHV G A L+K +HP+WSP+AIKSAIMTT
Sbjct: 677 AWSPVA-----------GNMFNILSGTSMACPHVTGIATLVKAVHPSWSPSAIKSAIMTT 823
Query: 589 VITYLSK 595
T L K
Sbjct: 824 A-TILDK 841
>TC78957 similar to GP|14091078|gb|AAK53589.1 subtilisin-like protein
{Glycine max}, partial (28%)
Length = 838
Score = 183 bits (465), Expect = 1e-46
Identities = 117/298 (39%), Positives = 162/298 (54%), Gaps = 26/298 (8%)
Frame = +1
Query: 9 LLVFYYYFFFLF---------------QCYIVYMGAHSHGPTPTSVDLETATSSHYDLLG 53
+L+F + FFFL Q YIVYMGA A S++ +L
Sbjct: 25 ILLFEFLFFFLGGESISSSATKSGNNNQVYIVYMGA--------------ANSTNAHVLN 162
Query: 54 SIVGSKEEAKEAIIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWE 113
+++ E+A ++++Y +GFAA L + EAA +A+ P VVSVF KLHTT SW+
Sbjct: 163 TVLRRNEKA---LVHNYKHGFSGFAARLSKNEAASIAQQPGVVSVFPDPILKLHTTHSWD 333
Query: 114 FLGLHGND------INSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRG-- 165
FL L + NS+ Q + +I +D+G+WPE+ SFSD G+ PIP+ W+G
Sbjct: 334 FLKLQTHVKIDSTLSNSSSQSS--SSDIVIGMLDSGIWPEATSFSDNGMDPIPSGWKGIC 507
Query: 166 ---GSSKKVPCNRKLIGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPG 222
CNRK+IGAR+ Y G + T RD VGHGTHT STA GN V G
Sbjct: 508 MTSNDFNSSNCNRKIIGARY----YPNLEGDDRVAA-TTRDTVGHGTHTASTAAGNAVSG 672
Query: 223 ASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVS 280
AS + + G KGGSP +R+A KVC ++ C G+ +L+A D AI DGVD++S+S
Sbjct: 673 ASYYGLAEGIAKGGSPESRLAI*KVCSNI----GCSGSAILAAFDDAISDGVDVLSLS 834
>BI307979 weakly similar to GP|20521377|db putative subtilisin-like protease
{Oryza sativa (japonica cultivar-group)}, partial (18%)
Length = 777
Score = 177 bits (449), Expect = 1e-44
Identities = 109/269 (40%), Positives = 154/269 (56%), Gaps = 1/269 (0%)
Frame = +3
Query: 263 LSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTP 322
L AID AI+DGVD+I++S G P+ E+ D ++ GA A+ +NI++V SAGN GP+P
Sbjct: 3 LKAIDDAIEDGVDVINLSIGFPAPLKYED---DVIAKGALQAVRKNIVVVCSAGNAGPSP 173
Query: 323 GSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFVDLPPNQSFTLVNSIDAKFS 381
S+ N +PW+ TV AST+DR F + I + + I G S+ N LV + D +++
Sbjct: 174 HSLSNPSPWIITVGASTVDRTFLAPIKLSNGTTIEGRSITPLRMGNSFCPLVLASDVEYA 353
Query: 382 NATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSG 441
+ ++ +C TLDPSKVKGKIV C R G+ V + E AG G+ L N KV
Sbjct: 354 GILSANSSYCLDNTLDPSKVKGKIVLCMR-GQGGRVKKSLEVQRAGGVGLILGNN-KVYA 527
Query: 442 NTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMAS 501
N + S+P+ + T G +T L + + T++ KPAP MA
Sbjct: 528 NDVPSDPYFIPTTG------VTYENTLKLVQYIHSSPNPMAQLLPGRTVLDTKPAPSMAI 689
Query: 502 FSSRGPNQVQPYILKPDVTAPGVNILAAY 530
FSSRGPN + P ILKPD+TAPGV+I AA+
Sbjct: 690 FSSRGPNIIDPNILKPDITAPGVDIFAAW 776
>TC82946 weakly similar to GP|20160863|dbj|BAB89802. putative
subtilisin-like protease {Oryza sativa (japonica
cultivar-group)}, partial (21%)
Length = 921
Score = 89.0 bits (219), Expect(2) = 2e-38
Identities = 51/133 (38%), Positives = 70/133 (52%)
Frame = +1
Query: 175 RKLIGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIK 234
R LIGA+FF+ + + + RD GHGTHT STA GN V AS F GT
Sbjct: 547 RNLIGAKFFNKGLQSKYPNITLGLNSTRDSHGHGTHTSSTAAGNRVDDASFFGYAPGTAS 726
Query: 235 GGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFT 294
G + +RVA YK W + +DV++AID AI DGVD++S+S G N ++
Sbjct: 727 GIASNSRVAMYKAIWDV----GVLSSDVIAAIDAAISDGVDVLSLSFG----INDVPLYD 882
Query: 295 DEVSIGAFHALAR 307
V+I F + +
Sbjct: 883 XPVAIATFAXMEK 921
Score = 89.0 bits (219), Expect(2) = 2e-38
Identities = 49/126 (38%), Positives = 65/126 (50%)
Frame = +3
Query: 52 LGSIVGSKEEAKEAIIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRS 111
L S+ SK +IY+Y +NGF+A L +E L +P +S KL TT S
Sbjct: 186 LNSLFSSK------LIYTYTHVMNGFSANLSPKEHESLKTSPGYISSIKDSHMKLDTTHS 347
Query: 112 WEFLGLHGNDINSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKV 171
+FLGL+ N AW FG + I+ IDTG+WPES SF D + IP+KW G +
Sbjct: 348 PQFLGLNPN--KGAWHDSNFGNDVIVGLIDTGIWPESESFKDNLMSEIPSKWNGQCENSI 521
Query: 172 PCNRKL 177
N L
Sbjct: 522 LFNSSL 539
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,654,481
Number of Sequences: 36976
Number of extensions: 241523
Number of successful extensions: 2096
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 1874
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1942
length of query: 609
length of database: 9,014,727
effective HSP length: 102
effective length of query: 507
effective length of database: 5,243,175
effective search space: 2658289725
effective search space used: 2658289725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133779.10


