

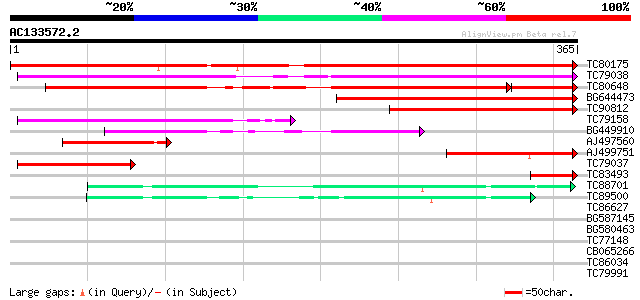
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.2 - phase: 0
(365 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80175 similar to GP|14582255|gb|AAK69429.1 ZIP-like zinc trans... 483 e-137
TC79038 similar to GP|15418778|gb|AAK37761.1 zinc transporter pr... 311 2e-85
TC80648 weakly similar to GP|15418778|gb|AAK37761.1 zinc transpo... 257 4e-77
BG644473 similar to PIR|T06385|T06 probable Fe(II) transport pro... 177 6e-45
TC90812 similar to GP|15418778|gb|AAK37761.1 zinc transporter pr... 144 7e-35
TC79158 similar to GP|15418778|gb|AAK37761.1 zinc transporter pr... 125 3e-29
BG449910 similar to PIR|T02478|T02 iron transport protein homolo... 117 9e-27
AJ497560 weakly similar to PIR|F86243|F862 ZIP4 probable zinc t... 105 3e-23
AJ499751 weakly similar to PIR|S33654|S336 zinc transport protei... 76 2e-14
TC79037 homologue to GP|15418778|gb|AAK37761.1 zinc transporter ... 65 4e-11
TC83493 similar to GP|12006859|gb|AAG44953.1 putative Zn transpo... 65 5e-11
TC88701 similar to GP|13162619|gb|AAG09635.1 zinc transporter {M... 60 1e-09
TC89500 zinc transporter [Medicago truncatula] 57 9e-09
TC86627 similar to PIR|T02334|T02334 probable urease accessory p... 34 0.10
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 33 0.23
BG580463 homologue to GP|15418778|gb| zinc transporter protein Z... 32 0.39
TC77148 ENOD20 31 0.86
CB065266 similar to GP|19033981|gb acetaldehyde dehydrogenase {P... 30 1.5
TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C5365... 30 1.9
TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity... 29 2.5
>TC80175 similar to GP|14582255|gb|AAK69429.1 ZIP-like zinc transporter
{Thlaspi caerulescens}, partial (69%)
Length = 1223
Score = 483 bits (1243), Expect = e-137
Identities = 255/377 (67%), Positives = 294/377 (77%), Gaps = 12/377 (3%)
Frame = +3
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVA 60
MTNS+C ESDLCRDE AA +LKF+AMASIL+AG +GIA+PL+G R LR+DG + A
Sbjct: 24 MTNSSCESGESDLCRDESAALILKFVAMASILVAGFSGIAVPLLGNRRGLLRSDGEILPA 203
Query: 61 AKAFAAGVILATGFVHMLSDATEALNSPCLPEFP--WSKFPFTGFFAMMAALFTLLLDFV 118
AKAFAAGVILATGFVHML DA +ALN CL + WS+FPFTGFFAMM+AL TLL+DFV
Sbjct: 204 AKAFAAGVILATGFVHMLQDAWKALNHSCLKSYSHVWSEFPFTGFFAMMSALLTLLVDFV 383
Query: 119 GTQYYERKQGMNRAVDEQARVGTSEEG--------NVGKVFGEEESGGMHIVGMHAHAAH 170
TQYYE + + D RV + EG + +V GE GGMHIVGMHAHA+
Sbjct: 384 ATQYYESQH--QKTHDRHGRVVGNGEGLEEELLGSGIVEVQGETFGGGMHIVGMHAHASQ 557
Query: 171 HRHNHP-HGDACDGGGIVKEHGHDHSHALIAANE-ETDVRHVVVSQVLELGIVSHSVIIG 228
H H+H HGD HGH HSH+ + ++ VRHVVVSQVLELGIVSHS+IIG
Sbjct: 558 HGHSHQNHGDG---------HGHGHSHSFGEHDGVDSSVRHVVVSQVLELGIVSHSLIIG 710
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVG 288
LSLGVSQSPC +RPLIAALSFHQFFEGFALGGCIS+A+FK SS TIMACFFALTTP+GV
Sbjct: 711 LSLGVSQSPCTMRPLIAALSFHQFFEGFALGGCISEARFKTSSATIMACFFALTTPLGVA 890
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IGT +AS +NPYSPGALI EGILD+LSAGILVYMALVDLIAADFLSK+M C+ RLQ+VS+
Sbjct: 891 IGTLVASNFNPYSPGALITEGILDSLSAGILVYMALVDLIAADFLSKKMRCSLRLQIVSF 1070
Query: 349 CMLFLGAGLMSSLAIWA 365
C+LFLGAG MSSLA+WA
Sbjct: 1071CLLFLGAGSMSSLALWA 1121
>TC79038 similar to GP|15418778|gb|AAK37761.1 zinc transporter protein ZIP1
{Glycine max}, partial (87%)
Length = 1546
Score = 311 bits (798), Expect = 2e-85
Identities = 175/360 (48%), Positives = 215/360 (59%)
Frame = +3
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E D RD A K A+ SIL+A G+ +PL+GK L + ++F KAFA
Sbjct: 129 CTCDEEDEERDRSKALRYKIAALVSILVASAIGVCLPLLGKVIPALSPEKDIFFIIKAFA 308
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVIL+TGF+H+L DA E L SPCL E PW FPFTGF AM A+ TL++D T Y++
Sbjct: 309 AGVILSTGFIHVLPDAFENLTSPCLNEHPWGDFPFTGFVAMCTAMGTLMVDTYATAYFQN 488
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
+++ E H H H H
Sbjct: 489 HYSKRAPAQVESQTTPDVENE-----------------------EHTHVHAHAS------ 581
Query: 186 IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIA 245
H H H H + E +RH V+SQVLELGI+ HSVIIG+SLG S+SP IRPL+A
Sbjct: 582 ----HSHAHGHIPFDQSSEL-LRHRVISQVLELGIIVHSVIIGISLGASESPKTIRPLVA 746
Query: 246 ALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGAL 305
AL+FHQFFEG LG CI+QA FK+ S TIM FFALTTP+G+GIG GI++VY+ SP AL
Sbjct: 747 ALTFHQFFEGMGLGSCITQANFKSLSITIMGLFFALTTPVGIGIGLGISNVYDENSPTAL 926
Query: 306 IAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
I EGI +A SAGIL+YMALVDL+AADF++ RM N RLQL S L LGAG MS +A WA
Sbjct: 927 IFEGIFNAASAGILIYMALVDLLAADFMNPRMQKNGRLQLGSNISLLLGAGCMSLIAKWA 1106
>TC80648 weakly similar to GP|15418778|gb|AAK37761.1 zinc transporter
protein ZIP1 {Glycine max}, partial (80%)
Length = 1330
Score = 257 bits (656), Expect(2) = 4e-77
Identities = 138/300 (46%), Positives = 186/300 (62%)
Frame = +1
Query: 24 KFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATE 83
K ++AS+L+ G G+++PL+ K L ++F KAFAAGVILATGF+H+L DA E
Sbjct: 184 KLGSIASVLVCGALGVSLPLLSKRIPILSPKNDIFFMIKAFAAGVILATGFIHILPDAFE 363
Query: 84 ALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRAVDEQARVGTSE 143
+LNSPCL E PW FP G AM++++ TL++D + YY+++ S+
Sbjct: 364 SLNSPCLKEKPWGDFPLAGLVAMLSSIATLMVDSFASSYYQKRH-----------FNPSK 510
Query: 144 EGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGHDHSHALIAANE 203
+ V +EE G H+ +H H H H H HG A +
Sbjct: 511 Q-----VPADEEKGDEHVGHVHVHT-HATHGHAHGSATSSQDSISPEL------------ 636
Query: 204 ETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCIS 263
+R ++SQVLELGIV HSVIIG+SLG +QS I+PL+ ALSFHQFFEG LGGCIS
Sbjct: 637 ---IRQRIISQVLELGIVVHSVIIGISLGTAQSIDTIKPLLVALSFHQFFEGMGLGGCIS 807
Query: 264 QAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMA 323
QA+F++ ST IMA FF+LTTPIG+ IG G++SVY SP +LI EG+ ++ SAGIL+YMA
Sbjct: 808 QAKFESRSTAIMATFFSLTTPIGIAIGMGVSSVYKDNSPTSLIVEGVFNSASAGILIYMA 987
Score = 49.3 bits (116), Expect(2) = 4e-77
Identities = 24/42 (57%), Positives = 31/42 (73%)
Frame = +2
Query: 324 LVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
LVDL+AADF+S RM NF++Q+ + L LG+G MS LA WA
Sbjct: 989 LVDLLAADFMSPRMQNNFKIQIGANISLLLGSGCMSLLAKWA 1114
>BG644473 similar to PIR|T06385|T06 probable Fe(II) transport protein root -
garden pea, partial (46%)
Length = 735
Score = 177 bits (449), Expect = 6e-45
Identities = 88/155 (56%), Positives = 115/155 (73%)
Frame = -3
Query: 211 VVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKAS 270
VV VLELGIV HSV+IGL +G S + C+I+ LIAA+ FHQ FEG LGGCI QA++K
Sbjct: 673 VVVMVLELGIVVHSVVIGLGMGASNNTCSIKGLIAAMCFHQMFEGMGLGGCILQAKYKFL 494
Query: 271 STTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAA 330
++ FF++TTP+G+ IG +++ Y SP ALI G+L+A SAG+L+YMALVDL+AA
Sbjct: 493 KNAMLVFFFSITTPLGIAIGLAMSTSYKENSPVALITVGLLNASSAGLLIYMALVDLLAA 314
Query: 331 DFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
DF+SKRM + +LQL SY +FLGAG MS +A WA
Sbjct: 313 DFMSKRMQSSIKLQLKSYVAVFLGAGGMSLMAKWA 209
>TC90812 similar to GP|15418778|gb|AAK37761.1 zinc transporter protein ZIP1
{Glycine max}, partial (34%)
Length = 829
Score = 144 bits (362), Expect = 7e-35
Identities = 74/121 (61%), Positives = 91/121 (75%)
Frame = +1
Query: 245 AALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGA 304
AAL+FHQFFEG LG CI+QA FK+ S TIM FFALTTP+G+ IG GI+S Y+ SP A
Sbjct: 13 AALTFHQFFEGMGLGSCITQANFKSLSITIMGLFFALTTPVGIAIGIGISSGYDENSPTA 192
Query: 305 LIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIW 364
LI EGI +A S+GIL+YMALVDL+AADF++ RM + L+L L LG+GLMS +A W
Sbjct: 193 LIVEGIFNAASSGILIYMALVDLLAADFMNPRMQKSGILRLGCNISLLLGSGLMSLIAKW 372
Query: 365 A 365
A
Sbjct: 373 A 375
>TC79158 similar to GP|15418778|gb|AAK37761.1 zinc transporter protein ZIP1
{Glycine max}, partial (38%)
Length = 752
Score = 125 bits (314), Expect = 3e-29
Identities = 73/179 (40%), Positives = 95/179 (52%)
Frame = +2
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DL RD+P A K A+ SIL+A G+ IPL+GK L + ++F KAFA
Sbjct: 215 CTCDEEDLDRDKPKALRYKIAALVSILVASGIGVCIPLLGKVIPALSPEKDIFFIIKAFA 394
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGF+H+L DA E L SP L + PW FPFTGF AM A+ TL++D T Y++
Sbjct: 395 AGVILATGFIHVLPDAFENLTSPRLKKHPWGDFPFTGFVAMCTAMGTLMVDTYATAYFQN 574
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGG 184
+ V E ++ G M + H HA+ H H HPH + G
Sbjct: 575 HYSKKAPAQVENEVSPDVE--------KDHEGHMDV---HTHAS-HGHAHPHMSSVSSG 715
>BG449910 similar to PIR|T02478|T02 iron transport protein homolog F23F1.1 -
Arabidopsis thaliana, partial (41%)
Length = 492
Score = 117 bits (292), Expect = 9e-27
Identities = 77/207 (37%), Positives = 103/207 (49%), Gaps = 1/207 (0%)
Frame = +2
Query: 62 KAFAAGVILATGFVHMLSDATEAL-NSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGT 120
K FAAGVIL+T VH+L DA AL + PW FPF+G ++ A+ L +D V +
Sbjct: 8 KCFAAGVILSTSLVHVLPDAYAALADCHVASRHPWKDFPFSGLVTLIGAILALFVDLVAS 187
Query: 121 QYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDA 180
+ E Q A VG E E GG GD
Sbjct: 188 SHVEHGQ--------YAPVGEKEM---------ELEGG------------------EGDC 262
Query: 181 CDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAI 240
G ++K ++ +VSQVLE+GI+ HSVIIG+++G+SQ+ C I
Sbjct: 263 ERGEELIK------------------LKQRLVSQVLEIGIIFHSVIIGVTMGMSQNVCTI 388
Query: 241 RPLIAALSFHQFFEGFALGGCISQAQF 267
RPL+AAL+FHQ FEG LGGC++QA F
Sbjct: 389 RPLVAALAFHQIFEGMGLGGCVAQAGF 469
>AJ497560 weakly similar to PIR|F86243|F862 ZIP4 probable zinc transporter
61460-62785 [imported] - Arabidopsis thaliana, partial
(15%)
Length = 630
Score = 105 bits (262), Expect = 3e-23
Identities = 53/70 (75%), Positives = 58/70 (82%)
Frame = +3
Query: 35 GMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNSPCLPEFP 94
G+ GI+IPLI H RYLRT+GNLFVA KAFA GVILA G VHML DA +ALNSPCLPEF
Sbjct: 3 GIVGISIPLILNHFRYLRTNGNLFVAMKAFAEGVILAKGCVHMLWDAIKALNSPCLPEF- 179
Query: 95 WSKFPFTGFF 104
W+KFPFTG F
Sbjct: 180 WTKFPFTGIF 209
>AJ499751 weakly similar to PIR|S33654|S336 zinc transport protein high
affinity - yeast (Saccharomyces cerevisiae), partial
(18%)
Length = 319
Score = 76.3 bits (186), Expect = 2e-14
Identities = 40/86 (46%), Positives = 57/86 (65%), Gaps = 2/86 (2%)
Frame = +1
Query: 282 TTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFL--SKRMSC 339
TTP G+ IG G+ Y+P S ALI +G+LD++SAGIL+Y ALV+LIA DF+ SK
Sbjct: 7 TTPFGIAIGLGVRETYDPDSQTALIVQGVLDSISAGILLYAALVELIANDFIYDSKFREK 186
Query: 340 NFRLQLVSYCMLFLGAGLMSSLAIWA 365
+ Q +++ L L AG+M+ + WA
Sbjct: 187 STIYQTLAFICLLL*AGIMALIGYWA 264
>TC79037 homologue to GP|15418778|gb|AAK37761.1 zinc transporter protein
ZIP1 {Glycine max}, partial (22%)
Length = 636
Score = 65.1 bits (157), Expect = 4e-11
Identities = 35/76 (46%), Positives = 46/76 (60%)
Frame = +1
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E D RD A K A+ SIL+A G+ +PL+GK L + ++F KAFA
Sbjct: 391 CTCDEEDEERDRSKALRYKIAALVSILVASAIGVCLPLLGKVIPALSPEKDIFFIIKAFA 570
Query: 66 AGVILATGFVHMLSDA 81
AGVIL+TGF+H+L DA
Sbjct: 571 AGVILSTGFIHVLPDA 618
>TC83493 similar to GP|12006859|gb|AAG44953.1 putative Zn transporter ZNT4
{Thlaspi caerulescens}, partial (7%)
Length = 176
Score = 64.7 bits (156), Expect = 5e-11
Identities = 30/30 (100%), Positives = 30/30 (100%)
Frame = +2
Query: 336 RMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
RMSCNFRLQLVSYCMLFLGAGLMSSLAIWA
Sbjct: 2 RMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 91
>TC88701 similar to GP|13162619|gb|AAG09635.1 zinc transporter {Medicago
truncatula}, partial (69%)
Length = 1235
Score = 60.5 bits (145), Expect = 1e-09
Identities = 68/316 (21%), Positives = 116/316 (36%), Gaps = 2/316 (0%)
Frame = +2
Query: 51 LRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAAL 110
LR + + FA GV L T +H LSDA E + ++PF A L
Sbjct: 257 LRWNEGFLILGTQFAGGVFLGTALMHFLSDANETFG-----DLTDKEYPFAYMLACAGYL 421
Query: 111 FTLLLDFVGTQYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAH 170
T+L D V + E+ A E V V + S G +
Sbjct: 422 ITMLADCVISSLLEKPNHGAGADVEGQGVDKGRSNGVNSQSQYQSSAGTN---------- 571
Query: 171 HRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLS 230
A +A + + V + + +HSV GL+
Sbjct: 572 -------------------------DADLAPSSSIGDTVYIFIYVYIIALCAHSVFEGLA 676
Query: 231 LGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQ--AQFKASSTTIMACFFALTTPIGVG 288
+GVS + + + H+ F A+G + + S A FA+++PIGV
Sbjct: 677 IGVSVTKADAWKALWTICLHKIFAAIAMGIALLRMVPNRPLLSCAAYAFAFAISSPIGVA 856
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IG + S + + A I L+ G+ +Y+++ L A ++ + S + +
Sbjct: 857 IGIVLDSTTQGHVADWIFA--ISMGLACGVFIYVSINHLFAKGYVPHKHS-KADSAYMKF 1027
Query: 349 CMLFLGAGLMSSLAIW 364
+ LG G+++ + IW
Sbjct: 1028LAVSLGIGVIAVVMIW 1075
>TC89500 zinc transporter [Medicago truncatula]
Length = 1040
Score = 57.4 bits (137), Expect = 9e-09
Identities = 70/291 (24%), Positives = 113/291 (38%), Gaps = 2/291 (0%)
Frame = +3
Query: 50 YLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAA 109
Y R + + FA GV L T +H LSD+ E + +PF A
Sbjct: 291 YFRWNEVFLLLGTQFAGGVFLGTSMMHFLSDSNETFE-----DLTKKTYPFAFMLACSGY 455
Query: 110 LFTLLLDFVGTQYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAA 169
L T+ D V Q +A+V E G +EE G
Sbjct: 456 LLTMFGDCVVVYVTSNNQ-------REAKVEELEGGRTP----QEEEG------------ 566
Query: 170 HHRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGL 229
E D S+ +A + T+V ++ L L + HSV G+
Sbjct: 567 -----------------TTELAMDESN--VAFMKTTNVGDTIL---LILALCFHSVFEGI 680
Query: 230 SLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKAS--STTIMACFFALTTPIGV 287
++G+S + + +S H+ F A+G + + K +T + FA+++PIGV
Sbjct: 681 AVGISGTKEEAWRNLWTISLHKIFAAIAMGIALLRMLPKRPLITTAGYSFAFAISSPIGV 860
Query: 288 GIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMS 338
GIG I + + + A I ++ G+ VY+A+ LI+ F +R S
Sbjct: 861 GIGIAIDATTEGKTADWMYA--ISMGIACGVFVYVAINHLISKGFKPQRKS 1007
>TC86627 similar to PIR|T02334|T02334 probable urease accessory protein
[imported] - Arabidopsis thaliana, partial (93%)
Length = 1246
Score = 33.9 bits (76), Expect = 0.10
Identities = 20/46 (43%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +3
Query: 156 SGGMHIVGMHAHAAHHR-HNHPHGDACDGGGIVKEHGHD----HSH 196
SGG H +H H HH H+H HGD+ G H HD HSH
Sbjct: 57 SGGDH-THVHDHDHHHHDHDHKHGDSFIGADGKVYHSHDGLAPHSH 191
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 32.7 bits (73), Expect = 0.23
Identities = 22/62 (35%), Positives = 26/62 (41%), Gaps = 22/62 (35%)
Frame = +1
Query: 146 NVGKVFGEEESGGMH-------IVGMHAHAAH--------HRHNHP-------HGDACDG 183
NVG++ E +SG MH I G+H H A HRH P G A G
Sbjct: 229 NVGRIHNETQSGQMHLWRHLRRIFGIHRHPARNRGES*ADHRHTRPP*SKE*QRGPATHG 408
Query: 184 GG 185
GG
Sbjct: 409 GG 414
>BG580463 homologue to GP|15418778|gb| zinc transporter protein ZIP1 {Glycine
max}, partial (5%)
Length = 283
Score = 32.0 bits (71), Expect = 0.39
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Frame = +2
Query: 170 HHRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVR-HVVVSQVLELGI 220
+ H H H A H H+H I+ ++ +++ H V+SQVLELGI
Sbjct: 158 NEEHTHVHAHA----------SHSHAHGHISFDQSSELLPHRVISQVLELGI 283
>TC77148 ENOD20
Length = 1108
Score = 30.8 bits (68), Expect = 0.86
Identities = 13/41 (31%), Positives = 19/41 (45%), Gaps = 3/41 (7%)
Frame = +1
Query: 159 MHIVGMHAHAAHHRHNHPHGDACDG---GGIVKEHGHDHSH 196
+H++ H H HH H+H H G + + H H H H
Sbjct: 499 LHLLHRHHHLHHHHHHHHHLGQLQFLILGNVHQLHHHHHHH 621
>CB065266 similar to GP|19033981|gb acetaldehyde dehydrogenase {Pseudomonas
putida}, partial (48%)
Length = 464
Score = 30.0 bits (66), Expect = 1.5
Identities = 22/70 (31%), Positives = 26/70 (36%), Gaps = 13/70 (18%)
Frame = +2
Query: 167 HAAHHRHNHPHGDACDGGGIVKEHGH------------DHSHALIAANEETDVRHVVVSQ 214
H H H HGD GG+ H DH HA++ V H V
Sbjct: 2 HLGHA*HRRDHGD----GGLATAGDHVDVSRLQLLLEVDHGHAVLTDGRRGQVDHAQVGL 169
Query: 215 VL-ELGIVSH 223
VL + GIV H
Sbjct: 170 VLAQEGIVRH 199
>TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C53655)
AU056546(S20671) AU056545(S20671) correspond to a region
of the predicted, partial (42%)
Length = 2412
Score = 29.6 bits (65), Expect = 1.9
Identities = 13/28 (46%), Positives = 14/28 (49%)
Frame = +3
Query: 158 GMHIVGMHAHAAHHRHNHPHGDACDGGG 185
G H H H HH H+H HG GGG
Sbjct: 1134 GGHYQQQHHHQPHHSHHHHHG----GGG 1205
>TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (8%)
Length = 1023
Score = 29.3 bits (64), Expect = 2.5
Identities = 11/32 (34%), Positives = 15/32 (46%)
Frame = -3
Query: 165 HAHAAHHRHNHPHGDACDGGGIVKEHGHDHSH 196
H H+ H HNHPH ++ H H+ H
Sbjct: 820 HHHSHTHHHNHPH---------IQHHNHNQHH 752
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,290,909
Number of Sequences: 36976
Number of extensions: 164271
Number of successful extensions: 1767
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1515
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1675
length of query: 365
length of database: 9,014,727
effective HSP length: 97
effective length of query: 268
effective length of database: 5,428,055
effective search space: 1454718740
effective search space used: 1454718740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC133572.2


