

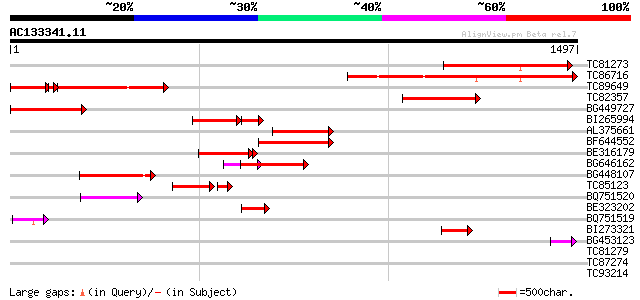
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133341.11 + phase: 0 /partial
(1497 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81273 similar to PIR|T14278|T14278 myosin-like protein my4 - c... 569 e-162
TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - c... 549 e-156
TC89649 similar to PIR|T14276|T14276 myosin-like protein my2 - c... 370 e-148
TC82357 similar to GP|7243765|gb|AAF43440.1| unconventional myos... 408 e-114
BG449727 similar to GP|7243765|gb unconventional myosin XI {Vall... 358 1e-98
BI265994 similar to PIR|F96587|F9 hypothetical protein T22H22.1 ... 213 3e-76
AL375661 weakly similar to PIR|S51824|S5 myosin heavy chain MYA2... 254 2e-67
BF644552 similar to PIR|T14279|T1 myosin-like protein my5 - comm... 213 4e-55
BE316179 similar to GP|15451591|g Putative myosin heavy chain {O... 203 4e-52
BG646162 similar to GP|15451591|g Putative myosin heavy chain {O... 201 1e-51
BG448107 similar to GP|11994771|d myosin-like protein {Arabidops... 171 1e-42
TC85123 homologue to PIR|G96539|G96539 hypothetical protein F14I... 140 8e-39
BQ751520 weakly similar to SP|P70569|MY5 Myosin Vb (Myosin 5B) (... 113 5e-25
BE323202 similar to PIR|S51823|S51 myosin heavy chain ATM2 - Ara... 86 1e-16
BQ751519 similar to GP|940860|emb|C MYO2 {Saccharomyces cerevisi... 69 1e-11
BI273321 similar to GP|20503048|gb putative myosin heavy chain {... 63 7e-10
BG453123 similar to PIR|D85390|D85 myosin-like protein [imported... 44 6e-04
TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.... 42 0.001
TC87274 similar to PIR|S51232|S51232 gibberellin-responsive ovar... 40 0.009
TC93214 similar to GP|10177918|dbj|BAB11329. kinesin-like protei... 39 0.020
>TC81273 similar to PIR|T14278|T14278 myosin-like protein my4 - common
sunflower, partial (22%)
Length = 1078
Score = 569 bits (1467), Expect = e-162
Identities = 287/353 (81%), Positives = 317/353 (89%), Gaps = 13/353 (3%)
Frame = +1
Query: 1145 WKSFEAERSSVFDRLIQMIGSAIEDQDDNALMAYWLSNTSALLFLLEQSLKTGTSTNATP 1204
WKSFEAER+SVFDRLIQMIGSAIE+QDDN LMAYWLSNTSALLFLL+QSLK+G ST++TP
Sbjct: 1 WKSFEAERTSVFDRLIQMIGSAIENQDDNDLMAYWLSNTSALLFLLQQSLKSGGSTDSTP 180
Query: 1205 NGKPPNPTSLFGRMTKSFLSSPSSANLASPSSVVRKVEAKYPALLFKQQLTAYLEKIYGI 1264
KPPNPTSLFGRMT F SSPSSANL +P VVRKV+AKYPALLFKQQLTAY+EKIYGI
Sbjct: 181 VRKPPNPTSLFGRMTMGFRSSPSSANLPAPLEVVRKVDAKYPALLFKQQLTAYVEKIYGI 360
Query: 1265 IRDNLTKELTSALALCIQAPRTSKGVLRSGRSFGKDSPMVHWQSIIESLNTLLCTLKENF 1324
+RDNL KEL S ++LCIQAPRTSKGVLRSGRSF KDSPM HWQSIIESLNTLLCT+KENF
Sbjct: 361 LRDNLKKELQSWISLCIQAPRTSKGVLRSGRSFSKDSPMGHWQSIIESLNTLLCTMKENF 540
Query: 1325 IPPVLIRKIFSQTFAFINVQLFN-----------SNGEYVKAGLAELELWCCQAKEEYAG 1373
+PPVLI+KIFSQTF++INVQLFN SNGEYVKAGLAELELWCCQAKEEYAG
Sbjct: 541 VPPVLIQKIFSQTFSYINVQLFNSLLLRRDCCTFSNGEYVKAGLAELELWCCQAKEEYAG 720
Query: 1374 SSWDELKHIRQAVGFLVIHQKYRISYDEIVNDLCPILSVQQLCKICTLYWDDNYNTRSVS 1433
+SWDELKHIRQAVGFLVIHQKYRISYDEI+NDLCPI+SVQQL ++CTLYWD NYNTR+VS
Sbjct: 721 TSWDELKHIRQAVGFLVIHQKYRISYDEIINDLCPIMSVQQLYRVCTLYWDANYNTRTVS 900
Query: 1434 PHVLASMR--MDLDSNDAMNDSFLLDDSSSIPFSVDDLSTSLQEKDFSDMKPA 1484
VL+SM+ M DSN+A +DSFLLDD+SSIPFSVDDLSTSLQE+D S+MKPA
Sbjct: 901 QDVLSSMKVLMAEDSNNAQSDSFLLDDTSSIPFSVDDLSTSLQERDCSEMKPA 1059
>TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - common
sunflower, partial (38%)
Length = 2675
Score = 549 bits (1414), Expect = e-156
Identities = 309/637 (48%), Positives = 428/637 (66%), Gaps = 32/637 (5%)
Frame = +2
Query: 893 KLEKQVEDLTLRLQLEKRLRVDVEEAKAKENERLQSALQKMQLQFKETKVLLEKEREATK 952
KLEK+VE+LT RLQ+EKRLR D+EE KA+E +L+ AL MQ+Q +E + KEREA++
Sbjct: 2 KLEKRVEELTWRLQIEKRLRTDLEEEKAQEVAKLRDALHAMQIQVEEANAKVIKEREASQ 181
Query: 953 K-LEARVPVIQEVPAVDHALLEKLSSENEKLKTLVSSL--EKKIDETEKRYEEEAKV-SE 1008
K ++ PVI+E P + EK++S ++ L SL E++ E KR + E + S+
Sbjct: 182 KAIQDAPPVIKETPVIIEDT-EKINSLLAEVNCLKESLLLEREAKEEAKRAQAETEARSK 358
Query: 1009 ERLKQALDAESKVIQMKTAMQRLEEKFADIEFANHVLQKQSLSINSPVKT-AVENLSTPV 1067
E K+ D++ K Q++ +QRLEEK ++ E N VL++Q+L+++ K+ A S +
Sbjct: 359 ELFKKVEDSDRKADQLQELVQRLEEKISNSESENQVLRQQALAVSPTAKSLAARPRSVII 538
Query: 1068 SEKLENGHHV---AEEPYDADTYVTPVKQFVAESDVKLKRSCSERHHGSFDSLVNCVSKN 1124
ENG+ + A+ P D ++ V++ ES+ K ++S +++ + D L+ C+S++
Sbjct: 539 QRTPENGNALNGEAKTPSDMTLALSNVRE--PESEGKPQKSLNDKQQENQDVLIKCISQD 712
Query: 1125 IGFNHGKPIAAFTIYKCLLHWKSFEAERSSVFDRLIQMIGSAIEDQDDNALMAYWLSNTS 1184
+GF+ GKPIAA IYKCLLHW+SFE ER++VFDR+IQ I SA+E QD+ ++AYWLSNTS
Sbjct: 713 LGFSEGKPIAACVIYKCLLHWRSFEVERTTVFDRIIQTIASAVEAQDNTDVLAYWLSNTS 892
Query: 1185 ALLFLLEQSLKTGTSTNATPNGKPPNPTSLFGRMTKSFLSSPSSANL-------ASPSSV 1237
LL LL+++LK + + TP + +SLFGRM++ SP SA L S
Sbjct: 893 TLLMLLQRTLKASGAASLTPQRRRTASSSLFGRMSQGLRGSPQSAGLPFINGRGLSRLDG 1072
Query: 1238 VRKVEAKYPALLFKQQLTAYLEKIYGIIRDNLTKELTSALALCIQAPRTSKGVLRSGRS- 1296
+R+VEAKYPALLFKQQLTA+LEK+YG+IRDNL KE++ L LCIQAPRTS+ L GRS
Sbjct: 1073 LRQVEAKYPALLFKQQLTAFLEKLYGMIRDNLKKEISPLLGLCIQAPRTSRQGLVKGRSH 1252
Query: 1297 ---FGKDSPMVHWQSIIESLNTLLCTLKENFIPPVLIRKIFSQTFAFINVQLFN------ 1347
+ + + HWQSI++SLN L +K N+ PP L+RK+F+Q F+FI+VQLFN
Sbjct: 1253 ANAVAQQALIAHWQSIVKSLNNSLKIMKANYAPPFLVRKVFTQIFSFIDVQLFNSLLLRR 1432
Query: 1348 -----SNGEYVKAGLAELELWCCQAKEEYAGSSWDELKHIRQAVGFLVIHQKYRISYDEI 1402
SNGEYVK GLAELE WC +A EEY GS+W+ELKHIRQAVGFLVIHQK + S +EI
Sbjct: 1433 ECCSFSNGEYVKTGLAELEQWCIEATEEYTGSAWEELKHIRQAVGFLVIHQKPKKSLNEI 1612
Query: 1403 VNDLCPILSVQQLCKICTLYWDDNYNTRSVSPHVLASMRMDL--DSNDAMNDSFLLDDSS 1460
+LCP LS+QQL +I T+YWDD Y T SVS +MR + DS +A++ SFLLDD S
Sbjct: 1613 TKELCPGLSIQQLYRISTMYWDDKYGTHSVSTEGTTTMRAMVAEDSTNAVSTSFLLDDDS 1792
Query: 1461 SIPFSVDDLSTSLQEKDFSDMKPADELLENPAFQFLI 1497
SIPFSVDD+S S+QE + +D+ P + EN F FL+
Sbjct: 1793 SIPFSVDDISKSMQEVEVADVDPPPLIRENSGFGFLL 1903
>TC89649 similar to PIR|T14276|T14276 myosin-like protein my2 - common
sunflower (fragment), partial (33%)
Length = 1356
Score = 370 bits (949), Expect(3) = e-148
Identities = 200/298 (67%), Positives = 228/298 (76%), Gaps = 5/298 (1%)
Frame = +3
Query: 126 ELSPHPFAVADAAYRLMINEGISQSILVSGESGAGKTETTKLLMRYLAYMGGRAAVAEGR 185
ELSPH FAVAD AYR MINEG S SILVSGESGAGKTETTK+LMRYLAY+GGR+ V EGR
Sbjct: 477 ELSPHVFAVADVAYRAMINEGKSNSILVSGESGAGKTETTKMLMRYLAYLGGRSGV-EGR 653
Query: 186 TVEQKVLESNPVLEAFGNAKTVRNNNSSRFGKFVEIQFDQKGRISGAAIRTYLLERSRVC 245
TVEQ+VLESNPVLEAFGNAKTVRNNNSSRFGKFVEIQFD KGRISGAAIRTYLLERSRVC
Sbjct: 654 TVEQQVLESNPVLEAFGNAKTVRNNNSSRFGKFVEIQFDNKGRISGAAIRTYLLERSRVC 833
Query: 246 QLSDPERNYHCFYMLCAAPAEVVKKYKLGHPRTFHYLNQSNCYELEGLDESKEYITIRRA 305
Q+SDPERNYHCFY+LCAAPAE +K+KLG+P +FHYLNQS CY L+G+ ++
Sbjct: 834 QVSDPERNYHCFYLLCAAPAEEREKFKLGNPSSFHYLNQSKCYLLDGVR*C*RISCNSKS 1013
Query: 306 MDVVGISIENQ--DAIFQVVAAILHLGNIEFVK---GDEIDSSMPKDEKSRFHLQTAAEL 360
NQ A Q + + +E + G+EIDSS+ KDE SRFHL T AEL
Sbjct: 1014NGCCW----NQ*GGARMQFLGS*QQSCTLEILNLQXGEEIDSSVVKDENSRFHLNTTAEL 1181
Query: 361 FMCDANALEDSLCKRVIVTRDETITKWLDPEAAALSRDALAKIVYTRLFDWLVDTINN 418
CDAN+LED+L +RV+VT E T+ LDP AA S DA AK +Y+RLFDWLV+ NN
Sbjct: 1182LKCDANSLEDALIQRVMVTPXEVXTRTLDPVAALSSXDAXAKTMYSRLFDWLVEKXNN 1355
Score = 152 bits (383), Expect(3) = e-148
Identities = 70/107 (65%), Positives = 84/107 (78%)
Frame = +1
Query: 1 AAVANPIVGTHVWIEDSDIAWIDGEVVGVNGEEIKVLCTSGKTVVVKASKIYHKDTEVPP 60
+A N IVG+HVW+ED +AWI GEV +NG E+ V GKTVV SK++ KD E PP
Sbjct: 100 SAPVNIIVGSHVWVEDPALAWIGGEVTKINGGEVHVRTGDGKTVVKSISKVFPKDNEAPP 279
Query: 61 SGVDDMTKLAYLHEPGVLNNLRSRYDINEIYTYTGNILIAVNPFIKL 107
GVDDMTKL+YLHEPGVLNNL +RY++NEIYTYTGNILIA+NPF K+
Sbjct: 280 GGVDDMTKLSYLHEPGVLNNLATRYELNEIYTYTGNILIAINPFSKV 420
Score = 44.7 bits (104), Expect(3) = e-148
Identities = 18/25 (72%), Positives = 20/25 (80%)
Frame = +2
Query: 103 PFIKLPHLYDIHMMAQYKGVAFGEL 127
PF +LPHLYD HMM QYKG AFG +
Sbjct: 407 PFQRLPHLYDTHMMQQYKGAAFGRV 481
>TC82357 similar to GP|7243765|gb|AAF43440.1| unconventional myosin XI
{Vallisneria gigantea}, partial (8%)
Length = 618
Score = 408 bits (1048), Expect = e-114
Identities = 204/205 (99%), Positives = 204/205 (99%)
Frame = +3
Query: 1037 DIEFANHVLQKQSLSINSPVKTAVENLSTPVSEKLENGHHVAEEPYDADTYVTPVKQFVA 1096
DIEFANHVLQKQSLSINSPVKTAVENLSTPVSEKLENGHHVAEEPYDADTYVTPVKQFVA
Sbjct: 3 DIEFANHVLQKQSLSINSPVKTAVENLSTPVSEKLENGHHVAEEPYDADTYVTPVKQFVA 182
Query: 1097 ESDVKLKRSCSERHHGSFDSLVNCVSKNIGFNHGKPIAAFTIYKCLLHWKSFEAERSSVF 1156
ESDVKLKRSCSE HHGSFDSLVNCVSKNIGFNHGKPIAAFTIYKCLLHWKSFEAERSSVF
Sbjct: 183 ESDVKLKRSCSELHHGSFDSLVNCVSKNIGFNHGKPIAAFTIYKCLLHWKSFEAERSSVF 362
Query: 1157 DRLIQMIGSAIEDQDDNALMAYWLSNTSALLFLLEQSLKTGTSTNATPNGKPPNPTSLFG 1216
DRLIQMIGSAIEDQDDNALMAYWLSNTSALLFLLEQSLKTGTSTNATPNGKPPNPTSLFG
Sbjct: 363 DRLIQMIGSAIEDQDDNALMAYWLSNTSALLFLLEQSLKTGTSTNATPNGKPPNPTSLFG 542
Query: 1217 RMTKSFLSSPSSANLASPSSVVRKV 1241
RMTKSFLSSPSSANLASPSSVVRKV
Sbjct: 543 RMTKSFLSSPSSANLASPSSVVRKV 617
>BG449727 similar to GP|7243765|gb unconventional myosin XI {Vallisneria
gigantea}, partial (12%)
Length = 613
Score = 358 bits (918), Expect = 1e-98
Identities = 176/202 (87%), Positives = 185/202 (91%), Gaps = 1/202 (0%)
Frame = +3
Query: 1 AAVANPIVGTHVWIEDSDIAWIDGEVVGVNGEEIKVLCTSGKTVVVKASKIYHKDTEVPP 60
A A P+VG VW+EDSD+AWIDGEV+ VNGE+IKVLCTSGKTV VKAS +YHKDTE PP
Sbjct: 6 ATAATPVVGAQVWVEDSDVAWIDGEVLEVNGEDIKVLCTSGKTVTVKASSVYHKDTEAPP 185
Query: 61 SGVDDMTKLAYLHEPGVLNNLRSRYDINEIYTYTGNILIAVNPFIKLPHLYDIHMMAQYK 120
GVDDMTKLAYLHEPGVL+NLRSRYDINEIYTYTGNILIAVNPFIKLPHLYD HMMAQYK
Sbjct: 186 CGVDDMTKLAYLHEPGVLDNLRSRYDINEIYTYTGNILIAVNPFIKLPHLYDSHMMAQYK 365
Query: 121 GVAFGELSPHPFAVADAAYRLMINEGISQSILVSGESGAGKTETTKLLMRYLAYMGGRAA 180
G FGELSPHPFA+ADAAYRLMINEGISQSILVSGESGAGKTE+TKLLMRYLAYMGGRAA
Sbjct: 366 GAGFGELSPHPFAIADAAYRLMINEGISQSILVSGESGAGKTESTKLLMRYLAYMGGRAA 545
Query: 181 -VAEGRTVEQKVLESNPVLEAF 201
AE RTVEQKVLESNPVL AF
Sbjct: 546 NAAEXRTVEQKVLESNPVLXAF 611
>BI265994 similar to PIR|F96587|F9 hypothetical protein T22H22.1 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 643
Score = 213 bits (541), Expect(2) = 3e-76
Identities = 101/129 (78%), Positives = 112/129 (86%)
Frame = +2
Query: 482 SYIEFVDNQDILDLIEKKPGGIISLLDEACMFPRSTHETFSQKLYQTFKDHKRFSKPKLS 541
SY+EFVDNQD+LDLIEKKPGGII+LLDEACMFP+STHETFS KLYQTFKDHKRF KPKL+
Sbjct: 2 SYLEFVDNQDVLDLIEKKPGGIIALLDEACMFPKSTHETFSTKLYQTFKDHKRFIKPKLA 181
Query: 542 PSDFTICHYAGDVTYQTEYFLDKNKDYVVAEHQSLLYASTCPFVSGLFPPSPEETSKQSK 601
SDF + HYAG V YQ+E F+DKNKDYVV EHQ +L S C FVSGLFPP EET+K +K
Sbjct: 182 RSDFNVVHYAGVVQYQSELFIDKNKDYVVPEHQVMLSTSKCSFVSGLFPPLSEETAKSAK 361
Query: 602 FSSIGSRFK 610
FSSIGSRFK
Sbjct: 362 FSSIGSRFK 388
Score = 92.8 bits (229), Expect(2) = 3e-76
Identities = 41/57 (71%), Positives = 49/57 (85%)
Frame = +1
Query: 612 QLQSLLETLSSTEPHYIRCVKPNNLLKPAIFDNKNVLLQLRCGGVMEAIRISCAGYP 668
QL+ L+E L+ TEPHYIRCVKPNNLLKP IF+N NV+ QLR GGV+EA+RI CAG+P
Sbjct: 418 QLKHLMEALNLTEPHYIRCVKPNNLLKPGIFENMNVMQQLRSGGVLEAVRIKCAGFP 588
>AL375661 weakly similar to PIR|S51824|S5 myosin heavy chain MYA2 -
Arabidopsis thaliana, partial (10%)
Length = 486
Score = 254 bits (649), Expect = 2e-67
Identities = 129/162 (79%), Positives = 148/162 (90%)
Frame = +1
Query: 694 EVTACKRILKNVWLEGYQIGKTKVFLRAGQMAELDTRRSEILGKSASIIQRKVRSYLARQ 753
EVTACKRIL+ V L+GYQIGKTKVFLRAGQMAELDT RSEILGKSASIIQRKVRSYLAR+
Sbjct: 1 EVTACKRILEKVGLKGYQIGKTKVFLRAGQMAELDTCRSEILGKSASIIQRKVRSYLARR 180
Query: 754 SFILLRVSALQIQAACRGQLARQVFEGMRREASSLLIQRCLRMHIAKKAYKELYASAVSI 813
SF+ +R+SA+Q+QAACRGQLARQV+EG+R+EASSL+IQRC RMHIA+KAY ELY+SA+SI
Sbjct: 181 SFVSIRLSAIQLQAACRGQLARQVYEGLRQEASSLIIQRCFRMHIARKAYTELYSSAISI 360
Query: 814 QTGMRVMAAHCELHSRRRTSAAIIIQSHCRKYLALLNFTKLK 855
QTGMR MAA CEL R++TSAAIIIQSHCR+YLA +F LK
Sbjct: 361 QTGMRGMAARCELRFRKQTSAAIIIQSHCRRYLAYHHFKNLK 486
>BF644552 similar to PIR|T14279|T1 myosin-like protein my5 - common
sunflower, partial (13%)
Length = 601
Score = 213 bits (542), Expect = 4e-55
Identities = 108/200 (54%), Positives = 154/200 (77%)
Frame = +2
Query: 656 VMEAIRISCAGYPTRKAFDEFVDRFGLLAPEVLDGSSEEVTACKRILKNVWLEGYQIGKT 715
V+EAIRISCAGYPTR+ F EF++RFG+LAPEVLDG+ ++ AC+ IL + ++GYQIGKT
Sbjct: 2 VLEAIRISCAGYPTRRTFYEFLNRFGVLAPEVLDGNYDDKVACQMILGKMGMKGYQIGKT 181
Query: 716 KVFLRAGQMAELDTRRSEILGKSASIIQRKVRSYLARQSFILLRVSALQIQAACRGQLAR 775
KVFLRAGQMAELD RRSE+LG +A IIQR+ R+++AR+ FI LR +A+ +Q+ RG LAR
Sbjct: 182 KVFLRAGQMAELDARRSEVLGNAARIIQRQTRTHIARKEFIELRRAAISLQSNLRGILAR 361
Query: 776 QVFEGMRREASSLLIQRCLRMHIAKKAYKELYASAVSIQTGMRVMAAHCELHSRRRTSAA 835
+++E +RREA++L I++ + +IA+K+Y + +SA +QTG+R M A E R++T AA
Sbjct: 362 KLYEKLRREAAALKIEKNFKGYIARKSYLKARSSATILQTGLRAMKARDEFXFRKQTKAA 541
Query: 836 IIIQSHCRKYLALLNFTKLK 855
I IQ+H R+ +A + L+
Sbjct: 542 IRIQAHFRRKIAYSYYKXLQ 601
Score = 40.4 bits (93), Expect = 0.005
Identities = 29/71 (40%), Positives = 41/71 (56%), Gaps = 3/71 (4%)
Frame = +2
Query: 825 ELHSRRRT---SAAIIIQSHCRKYLALLNFTKLKKAAIATQCAWRGKVARRELRKLKMAA 881
EL +RR +AA IIQ R ++A F +L++AAI+ Q RG +AR+ KL+
Sbjct: 212 ELDARRSEVLGNAARIIQRQTRTHIARKEFIELRRAAISLQSNLRGILARKLYEKLR--- 382
Query: 882 RETGALQDAKN 892
RE AL+ KN
Sbjct: 383 REAAALKIEKN 415
>BE316179 similar to GP|15451591|g Putative myosin heavy chain {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (9%)
Length = 478
Score = 203 bits (516), Expect(2) = 4e-52
Identities = 96/146 (65%), Positives = 118/146 (80%), Gaps = 1/146 (0%)
Frame = +2
Query: 498 KKPGGIISLLDEACMFPRSTHETFSQKLYQTFKDHKRFSKPKLSPSDFTICHYAGDVTYQ 557
+ P G+I+LLDEACMFP+STHETFS KL+Q F+ H R + + S +DF I HYAG VTY
Sbjct: 2 ENPIGVIALLDEACMFPKSTHETFSTKLFQNFRSHPRLASERFSQTDFIISHYAGKVTYH 181
Query: 558 TEYFLDKNKDYVVAEHQSLLYASTCPFVSGLFPPSPEETSKQS-KFSSIGSRFKQQLQSL 616
T+ FLDKN+DYVV EH +LL +S CPFVSGLFP PEE+S+ S KFSS+ +RFKQQLQ+L
Sbjct: 182 TDAFLDKNRDYVVVEHCNLLSSSNCPFVSGLFPLLPEESSRSSYKFSSVATRFKQQLQAL 361
Query: 617 LETLSSTEPHYIRCVKPNNLLKPAIF 642
+ETL STEPHYIRCVKPN+L +P +F
Sbjct: 362 METLKSTEPHYIRCVKPNSLNRPQMF 439
Score = 21.9 bits (45), Expect(2) = 4e-52
Identities = 7/13 (53%), Positives = 11/13 (83%)
Frame = +3
Query: 642 FDNKNVLLQLRCG 654
F+N +++ QLRCG
Sbjct: 438 FENASIIHQLRCG 476
>BG646162 similar to GP|15451591|g Putative myosin heavy chain {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (12%)
Length = 716
Score = 201 bits (512), Expect = 1e-51
Identities = 98/180 (54%), Positives = 135/180 (74%)
Frame = +3
Query: 609 FKQQLQSLLETLSSTEPHYIRCVKPNNLLKPAIFDNKNVLLQLRCGGVMEAIRISCAGYP 668
FKQQLQ+L+ETL +TEPHYIRCVKPN+ P F+N +VL QLRCGGV+EA+RIS AGYP
Sbjct: 174 FKQQLQALMETLKTTEPHYIRCVKPNSSNLPQKFENTSVLHQLRCGGVLEAVRISLAGYP 353
Query: 669 TRKAFDEFVDRFGLLAPEVLDGSSEEVTACKRILKNVWLEGYQIGKTKVFLRAGQMAELD 728
TR+ + EFVDRFGL+APE +DGS ++ ++IL+ + LE +Q+G+TKVFLRAGQ+ LD
Sbjct: 354 TRRTYSEFVDRFGLIAPEFMDGSYDDRATTQKILQKLKLENFQLGRTKVFLRAGQIGILD 533
Query: 729 TRRSEILGKSASIIQRKVRSYLARQSFILLRVSALQIQAACRGQLARQVFEGMRREASSL 788
+RRSE+L +A IQR++R+++A + F L A CRG LAR+++ R A+++
Sbjct: 534 SRRSEVLDNAAKFIQRRLRTFIAHRGFHLNPSCCCFS*ACCRGCLARKIYASKRETAAAI 713
Score = 64.7 bits (156), Expect = 3e-10
Identities = 42/106 (39%), Positives = 63/106 (58%), Gaps = 4/106 (3%)
Frame = +1
Query: 566 KDYVVAEHQSLLYASTCPFVSGLFPPSPEETSKQS-KFSSIGSRFKQQLQ---SLLETLS 621
+DYVV EH ++L +S CPFVS LFP PEE+S+ S KFSS+ SRFK + +L LS
Sbjct: 1 RDYVVLEHCNVLSSSKCPFVSSLFPSLPEESSRSSYKFSSVASRFKVYISITFRVLLNLS 180
Query: 622 STEPHYIRCVKPNNLLKPAIFDNKNVLLQLRCGGVMEAIRISCAGY 667
+ H + ++ +L+ IFD + +L+ + I++SC Y
Sbjct: 181 NNFKHLWKHLRQQSLI---IFDVSSQILRTYHKSL--KIQVSCINY 303
>BG448107 similar to GP|11994771|d myosin-like protein {Arabidopsis
thaliana}, partial (17%)
Length = 599
Score = 171 bits (434), Expect = 1e-42
Identities = 89/202 (44%), Positives = 134/202 (66%), Gaps = 1/202 (0%)
Frame = +1
Query: 184 GRTVEQKVLESNPVLEAFGNAKTVRNNNSSRFGKFVEIQFDQKGRISGAAIRTYLLERSR 243
G +E ++L++NP+LEAFGN KT+RN+NSSRFGK +EI F + G+ISGA I+T+LLE+SR
Sbjct: 1 GSGIEHEILKTNPILEAFGNGKTLRNDNSSRFGKLIEIHFSETGKISGANIQTFLLEKSR 180
Query: 244 VCQLSDPERNYHCFYMLCA-APAEVVKKYKLGHPRTFHYLNQSNCYELEGLDESKEYITI 302
V Q ++ ER+YH FY LCA AP+ + +K L + YL QSNCY + +D+++E+ +
Sbjct: 181 VVQCNEGERSYHIFYQLCAGAPSSLREKLNLRSVEDYKYLRQSNCYSINDVDDAEEFRIV 360
Query: 303 RRAMDVVGISIENQDAIFQVVAAILHLGNIEFVKGDEIDSSMPKDEKSRFHLQTAAELFM 362
A+DVV IS E+Q+ +F ++AA+L LGNI F D + +++ F + A+L
Sbjct: 361 TDALDVVHISKEDQENVFAMLAAVLWLGNISFTVIDNENHVQAVEDEGLF---STAKLIG 531
Query: 363 CDANALEDSLCKRVIVTRDETI 384
CD L+ +L R + +TI
Sbjct: 532 CDIEDLKLTLSTRKMKVGKDTI 597
>TC85123 homologue to PIR|G96539|G96539 hypothetical protein F14I3.6
[imported] - Arabidopsis thaliana, partial (14%)
Length = 544
Score = 140 bits (354), Expect(2) = 8e-39
Identities = 65/111 (58%), Positives = 82/111 (73%)
Frame = +3
Query: 430 IGVLDIYGFESFKNNSFEQFCINLTNEKLQQHFNQHVFKMEQEEYKKEEIDWSYIEFVDN 489
I +LDIYGFESF NSFEQFCIN NE+LQQHFN+H+FK+EQEEY ++ IDW+ +EF DN
Sbjct: 60 ISILDIYGFESFNRNSFEQFCINYANERLQQHFNRHLFKLEQEEYIQDGIDWAKVEFEDN 239
Query: 490 QDILDLIEKKPGGIISLLDEACMFPRSTHETFSQKLYQTFKDHKRFSKPKL 540
QD L+L EKKP G++SLLDE FP T TF+ KL Q + F + ++
Sbjct: 240 QDCLNLFEKKPLGLLSLLDEESTFPNGTDLTFANKLKQHLNSNSCFKEERV 392
Score = 39.7 bits (91), Expect(2) = 8e-39
Identities = 18/40 (45%), Positives = 25/40 (62%)
Frame = +2
Query: 549 HYAGDVTYQTEYFLDKNKDYVVAEHQSLLYASTCPFVSGL 588
HYAG+VTY T FL+KN+D + + LL +S C S +
Sbjct: 413 HYAGEVTYDTTAFLEKNRDLMHVDSIQLLSSSKCHLPSDI 532
>BQ751520 weakly similar to SP|P70569|MY5 Myosin Vb (Myosin 5B) (Myosin heavy
chain myr 6). [Rat] {Rattus norvegicus}, partial (7%)
Length = 549
Score = 113 bits (283), Expect = 5e-25
Identities = 63/164 (38%), Positives = 97/164 (58%), Gaps = 1/164 (0%)
Frame = -1
Query: 188 EQKVLESNPVLEAFGNAKTVRNNNSSRFGKFVEIQFDQKGRISGAAIRTYLLERSRVCQL 247
E+++L + P+++ K +SSRFGK++EI FD+K I GA IRTYLLERSR+
Sbjct: 525 EEQILATTPIMK-LRKRKDDAQ*HSSRFGKYIEIMFDEKTNIIGAKIRTYLLERSRLVFQ 349
Query: 248 SDPERNYHCFYMLCAAPAEVVK-KYKLGHPRTFHYLNQSNCYELEGLDESKEYITIRRAM 306
ERNYH FY L A ++ + + L F YLNQ NC ++G+D+ E+ + ++
Sbjct: 348 PLKERNYHIFYQLVAGASDKERQELSLLPIEEFEYLNQGNCPTIDGVDDKSEFEATKGSL 169
Query: 307 DVVGISIENQDAIFQVVAAILHLGNIEFVKGDEIDSSMPKDEKS 350
+G+S Q IF+++A +LHLGN++ + DS + E S
Sbjct: 168 KTIGVSEAQQTEIFKLLAGLLHLGNVK-IGASRNDSVLAPTEPS 40
>BE323202 similar to PIR|S51823|S51 myosin heavy chain ATM2 - Arabidopsis
thaliana (fragment), partial (7%)
Length = 354
Score = 85.9 bits (211), Expect = 1e-16
Identities = 41/75 (54%), Positives = 52/75 (68%)
Frame = +2
Query: 612 QLQSLLETLSSTEPHYIRCVKPNNLLKPAIFDNKNVLLQLRCGGVMEAIRISCAGYPTRK 671
QL L+ L ST PH+IRC+KPN P I+DN+ VL QLRC G++EA+RIS AGYPTR
Sbjct: 110 QLFKLMHQLESTTPHFIRCIKPNTKKLPGIYDNELVLQQLRCCGLLEAVRISRAGYPTRI 289
Query: 672 AFDEFVDRFGLLAPE 686
+F R+G+L E
Sbjct: 290 KHQDFSRRYGILLSE 334
>BQ751519 similar to GP|940860|emb|C MYO2 {Saccharomyces cerevisiae}, partial
(7%)
Length = 794
Score = 69.3 bits (168), Expect = 1e-11
Identities = 43/108 (39%), Positives = 58/108 (52%), Gaps = 13/108 (12%)
Frame = +2
Query: 8 VGTHVWIEDSDIAWIDGEVVGVNGEEIKVLCT----SG--KTVVVKASKIYHKDTEVPP- 60
VGT W D+ W+ EVV E KV SG KT+ V A+ + + D+ +PP
Sbjct: 428 VGTRAWQPDATEGWVASEVVNKTEEGNKVKLVFKLESGEEKTIEVTAAALQNGDSSLPPL 607
Query: 61 ------SGVDDMTKLAYLHEPGVLNNLRSRYDINEIYTYTGNILIAVN 102
DD+T L++L+EP VL +R RY EIYTY+G +LIA N
Sbjct: 608 MNPTMLEASDDLTNLSHLNEPAVLQAIRLRYAQKEIYTYSGIVLIAAN 751
>BI273321 similar to GP|20503048|gb putative myosin heavy chain {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 294
Score = 63.2 bits (152), Expect = 7e-10
Identities = 34/85 (40%), Positives = 52/85 (61%), Gaps = 2/85 (2%)
Frame = +1
Query: 1139 YKCLLHWKSFEAERSSVFDRLIQMIGSAIEDQDDNALMAYWLSNTSALLFLLEQSLKTG- 1197
YKCLLHW +FE+ER+++FD +I I I+ +DD+ ++ LSNTSAL LL++++++
Sbjct: 16 YKCLLHWHAFESERTAIFDYIIDGINEVIKVRDDDIVLPXXLSNTSALXCLLQRNVRSNG 195
Query: 1198 -TSTNATPNGKPPNPTSLFGRMTKS 1221
+T A TS G KS
Sbjct: 196 FLTTTAQRYAGSSGLTSRIGHGLKS 270
>BG453123 similar to PIR|D85390|D85 myosin-like protein [imported] -
Arabidopsis thaliana, partial (5%)
Length = 642
Score = 43.5 bits (101), Expect = 6e-04
Identities = 26/72 (36%), Positives = 42/72 (58%), Gaps = 2/72 (2%)
Frame = +2
Query: 1427 YNTRSVSPHVLASMR--MDLDSNDAMNDSFLLDDSSSIPFSVDDLSTSLQEKDFSDMKPA 1484
Y T+SVS V++ MR ++ D+ + ++SFLLDD SIPFS +D+ ++ D ++
Sbjct: 2 YGTQSVSNEVVSVMREMVNKDNQNMPSNSFLLDDDLSIPFSAEDVDMAIPPIDLDEIDLP 181
Query: 1485 DELLENPAFQFL 1496
+ E QFL
Sbjct: 182 LFVSEYSCAQFL 217
>TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.170 -
Arabidopsis thaliana, partial (31%)
Length = 1205
Score = 42.4 bits (98), Expect = 0.001
Identities = 52/258 (20%), Positives = 106/258 (40%), Gaps = 23/258 (8%)
Frame = +2
Query: 868 KVARRELRKLKMAARETGALQDAKNKLEKQVEDLTLRLQLEK-RLRVDVEEAKAKE---- 922
++ R +L + ++E D+ +L + +E L++EK L V+ + A +
Sbjct: 185 EIKRLQLELTRQRSKEASNAMDSSKRLIETLEKENTTLKMEKSELEAAVKASSASDLSDP 364
Query: 923 ------NERLQSALQKMQLQFKETKVLLEKEREATKKLEARVPVIQEVPAVDHALLEKLS 976
E ++ +LQKM K+T+ + R +QE+ + LLEK +
Sbjct: 365 SKSFPGKEDMEISLQKMSNDLKKTQ-------------QERDKAVQELTRLKQHLLEKEN 505
Query: 977 SENEKLKTLVSSLEKKIDETE---------KRYEEEAKVSEERLKQALDAESKVIQMKTA 1027
E+EK+ +E+ D +R E+A +E+LK A ++E ++ +
Sbjct: 506 EESEKMDEDTKVIEELRDSNNYLRAQISHLERALEQATSDQEKLKSANNSE--ILTSREV 679
Query: 1028 MQRLEEKFADIEFANHVLQKQSLSINSPVKTAVENLSTPVSE---KLENGHHVAEEPYDA 1084
+ L +K + +S + NL T + + ++E H+ EE A
Sbjct: 680 IDDLNKKLTN-----------CISTIDAKNIELINLQTALGQYYAEIEAKEHLEEELARA 826
Query: 1085 DTYVTPVKQFVAESDVKL 1102
+ Q + ++D ++
Sbjct: 827 REETANLSQLLKDADSRV 880
>TC87274 similar to PIR|S51232|S51232 gibberellin-responsive ovarian protein
G14 - garden pea (fragment), partial (45%)
Length = 734
Score = 39.7 bits (91), Expect = 0.009
Identities = 35/135 (25%), Positives = 66/135 (47%), Gaps = 4/135 (2%)
Frame = +1
Query: 927 QSALQKMQLQFKETKVLLEKEREATKKLEARVPVIQEVPAVDHALLEK-LSSENEKLKTL 985
Q A +++ Q T V EKE K+E P EVPA + A E+ ++ ENE+ +
Sbjct: 73 QMATVEVETQQAPTTVT-EKETVEVTKVEETTPPASEVPASEPATTEEVVAVENEETEVP 249
Query: 986 VSSLEKKIDETEKRYEEEAKVSEERLKQALDAE--SKVIQMKTAMQRLEEK-FADIEFAN 1042
V + ++ + K E + + +K+ + AE + + ++A +++E K E A
Sbjct: 250 VEAESTQVTQEAKPEVENPEPEKVEVKEEVSAEEAKETTETESAPEKVEAKEEVSTEEAK 429
Query: 1043 HVLQKQSLSINSPVK 1057
+ +S +I +PV+
Sbjct: 430 ETTEIESAAIAAPVE 474
>TC93214 similar to GP|10177918|dbj|BAB11329. kinesin-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 715
Score = 38.5 bits (88), Expect = 0.020
Identities = 41/167 (24%), Positives = 68/167 (40%), Gaps = 24/167 (14%)
Frame = +3
Query: 875 RKLKMAARETGALQDAKNKLEKQVEDLTLRLQLEKRLRVDVEEAKAKENERLQSALQKMQ 934
RK+ E +Q ++ L +VE+L + + D K K E L+K Q
Sbjct: 33 RKITELEDEKRTVQQERDCLLAEVENLAANSDGQTQKLEDTHSQKLKALEAQILDLKKKQ 212
Query: 935 LQFKETKVLLEKEREATKKLEARVPVIQEVPAVDHALLEKLSSENEKLKTLVSSLEKKID 994
+ +K EATK+L+ IQ + A L +K+ E E+ + +S EK++
Sbjct: 213 ESQVQLMKQKQKSDEATKRLQDE---IQSIKAQKVQLQQKIKQEAEQFRQWKASREKELL 383
Query: 995 ETEK------------------------RYEEEAKVSEERLKQALDA 1017
+ K R EEA ++ +RLK+ L+A
Sbjct: 384 QLRKEGRKSEYEKHKLQALNQRQRMVLQRKTEEAAMATKRLKELLEA 524
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,262,780
Number of Sequences: 36976
Number of extensions: 576803
Number of successful extensions: 3116
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 3034
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3091
length of query: 1497
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1388
effective length of database: 4,984,343
effective search space: 6918268084
effective search space used: 6918268084
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC133341.11


