

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.5 + phase: 0 /pseudo
(1454 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
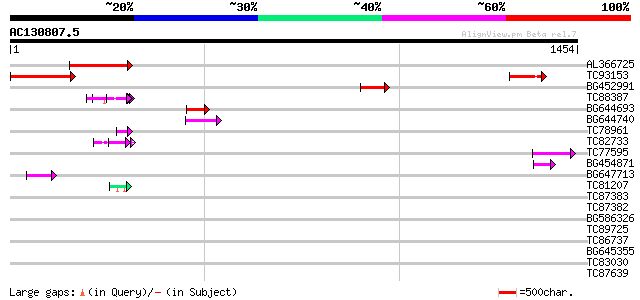
Score E
Sequences producing significant alignments: (bits) Value
AL366725 268 9e-72
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 268 9e-72
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 60 5e-09
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 54 6e-07
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 52 1e-06
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 52 1e-06
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 48 3e-05
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 47 4e-05
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 45 2e-04
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 45 2e-04
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 45 3e-04
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 43 8e-04
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 42 0.001
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 40 0.009
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 38 0.025
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 36 0.12
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 35 0.16
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 34 0.47
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 34 0.47
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 32 1.4
>AL366725
Length = 485
Score = 268 bits (686), Expect = 9e-72
Identities = 121/161 (75%), Positives = 141/161 (87%)
Frame = +2
Query: 153 KFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYKVM 212
KFYPHY AETAEFSKCIKFENGLR +IKRAIGYQQ+R+F DLV+ CRI EEDTKAH KV+
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 213 SERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDE 272
+ER+ KG+ SRPKPYSAPADKGKQR+ D+RRP ++DA AEIVCF GEKGHKSNVC K+
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEI 361
Query: 273 KKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
KKC RC +KGH++ADCKR D+VC+NCNEEGHI +QC QPK+
Sbjct: 362 KKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 268 bits (686), Expect = 9e-72
Identities = 130/171 (76%), Positives = 145/171 (84%), Gaps = 2/171 (1%)
Frame = +2
Query: 1 MAGRNDAAIAGALEAIAQAVEQQPQASNGE--VRMLETFLRNHPPAFKGMYDPDGAQSWL 58
M G +DAA+ ALEA+AQAV+Q P+ G RMLETFLRNHPP FKG Y PDGA WL
Sbjct: 2 MTGSSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWL 181
Query: 59 KEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNRY 118
KE+ERIFR MQC E +KV+FGTHMLAEEADDWW+SLLPVLEQD+AVVTWA+FR+EFL RY
Sbjct: 182 KEIERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRY 361
Query: 119 FLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCI 169
F EDV G+KEIEFLELK GD+SV EYAAKFVELA FYPHY+AETAEFSKCI
Sbjct: 362 FPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELATFYPHYSAETAEFSKCI 514
Score = 91.3 bits (225), Expect = 3e-18
Identities = 52/99 (52%), Positives = 62/99 (62%), Gaps = 2/99 (2%)
Frame = +2
Query: 1281 TDAAIAAALEAIAQAVGQQPQAGNGE--VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVE 1338
+DAA+ AALEA+AQAV Q P+ G RMLETFLRNHPP FKG Y PDGA WLKE+E
Sbjct: 14 SDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWLKEIE 193
Query: 1339 RIFRTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIE 1377
RIFR E Q ++ + E W S LP++E
Sbjct: 194 RIFRVMQCFE--TQKVQFGTHMLAEEADDWWISLLPVLE 304
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 60.5 bits (145), Expect = 5e-09
Identities = 30/74 (40%), Positives = 46/74 (61%)
Frame = +1
Query: 901 ICWNSLET*VWYVSCRLKAYSWAC*RLIVISWVVSEKHSKWMSSWLI**LLVMARRIVTS 960
+ NS ET*VW+V CR + +W C*R W+VS++ +WM SW I* L ++ ++V
Sbjct: 64 VALNSSET*VWFVKCRHRV*NWGC*RSTTNFWIVSKRLRRWM*SW*I*CLGIIRLKMVIL 243
Query: 961 RLMIREY*DSEVEF 974
+LMI+E +E+
Sbjct: 244 KLMIKECCSFGIEY 285
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 53.5 bits (127), Expect = 6e-07
Identities = 33/117 (28%), Positives = 48/117 (40%), Gaps = 9/117 (7%)
Frame = +1
Query: 212 MSERRGKGRQSRPKPYSAPADKGKQRLNDER---------RPNRRDASAEIVCFKCGEKG 262
+S R K R P S +R+ ER R +RR S + +C C G
Sbjct: 268 VSRSRSKSRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPG 447
Query: 263 HKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGK 319
H C + C C GH+ ++C + C+NC E GH+++ C T GK
Sbjct: 448 HYVRECP-NVAVCHNCSLPGHIASECSTKSL-CWNCKEPGHMASSCPNEGICHTCGK 612
Score = 52.8 bits (125), Expect = 1e-06
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Frame = +1
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCK-----RGDV-VCYNCNEE 301
+ S + +C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC ++
Sbjct: 517 ECSTKSLCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQ 693
Query: 302 GHISTQCTQPK 312
GHI+ +CT K
Sbjct: 694 GHIAVECTNEK 726
Score = 52.4 bits (124), Expect = 1e-06
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 5/123 (4%)
Frame = +1
Query: 196 SRCRICEEDTKAHYKVMSERRGKGRQSRPKPYSAPADKGKQRLN---DERRPNR--RDAS 250
SR R + +++ V R + R PY + +G + N + +RP R+
Sbjct: 289 SRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGHYVRECP 468
Query: 251 AEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQ 310
VC C GH ++ C+ + C+ C + GH+ + C + +C+ C + GH + +CT
Sbjct: 469 NVAVCHNCSLPGHIASECST-KSLCWNCKEPGHMASSCPN-EGICHTCGKAGHRARECTV 642
Query: 311 PKK 313
P+K
Sbjct: 643 PQK 651
Score = 38.1 bits (87), Expect = 0.025
Identities = 22/85 (25%), Positives = 31/85 (35%), Gaps = 23/85 (27%)
Frame = +1
Query: 247 RDASAEIVCFKCGEKGHKSNVCTKD----------------------EKKCFRCGQKGHL 284
RD + +C C GH + C K + C C Q GH+
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 285 LADCKRGDV-VCYNCNEEGHISTQC 308
DC G + +C NC GH + +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 36.2 bits (82), Expect = 0.095
Identities = 26/96 (27%), Positives = 35/96 (36%), Gaps = 41/96 (42%)
Frame = +1
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCK------------------------ 289
+C C ++GH + CT +EK C C + GHL DC
Sbjct: 670 LCNNCYKQGHIAVECT-NEKACNNCRKTGHLARDCPNDPICNLCNISGHVARQCPKSNVI 846
Query: 290 ---------RG--------DVVCYNCNEEGHISTQC 308
RG DVVC +C + GH+S C
Sbjct: 847 GDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHMSRDC 954
Score = 31.6 bits (70), Expect = 2.4
Identities = 15/44 (34%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Frame = +1
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK-CFRCGQKGHLLADCKRGDVV 294
++VC C + GH S C C CG +GH +C G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 52.4 bits (124), Expect = 1e-06
Identities = 24/59 (40%), Positives = 40/59 (67%)
Frame = +2
Query: 454 DEIPDVPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNVSP 512
D + VPPE +++F IDL+P P+ + YR++ +L LK QL+D L+K F++P++ P
Sbjct: 2 DHLL*VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP 178
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 52.4 bits (124), Expect = 1e-06
Identities = 36/93 (38%), Positives = 48/93 (50%)
Frame = -2
Query: 451 VFPDEIPDVPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNV 510
VFPD P +P ERE+ F IDL+ T+ +S P M +EL +LK L+D L+K F
Sbjct: 429 VFPDNFPVIPLEREIFFCIDLLLDTQLISNPP*LMDRTELKKLKI*LKDSLEKGFNNQVF 250
Query: 511 SPWERLCCW*RRRTVA*GCALTIVS*TKLQ*RT 543
CC +R GCAL + + Q RT
Sbjct: 249 LHRVLRCCLCEKRMGTLGCALITDNLIRSQQRT 151
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 47.8 bits (112), Expect = 3e-05
Identities = 22/43 (51%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +2
Query: 274 KCFRCGQKGHLLADCKRGD--VVCYNCNEEGHISTQC-TQPKK 313
+CF CG GH DCK GD CY C E GHI C PKK
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKK 484
Score = 42.4 bits (98), Expect = 0.001
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Frame = +2
Query: 255 CFKCGEKGHKSNVCTKDE--KKCFRCGQKGHLLADCK 289
CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 359 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 469
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 47.4 bits (111), Expect = 4e-05
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 12/67 (17%)
Frame = +3
Query: 254 VCFKCGEKGHKSNVCT-----KDEKKCFRCGQKGHLLADC----KRGDVV---CYNCNEE 301
+C +C +GH++ C +D K + CG GH LA+C + G + C+ C E+
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 302 GHISTQC 308
GH+S C
Sbjct: 534 GHLSKNC 554
Score = 46.2 bits (108), Expect = 9e-05
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 10/119 (8%)
Frame = +3
Query: 214 ERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKD-- 271
+++ K ++ +P S P GK+ L R P + + CF C H + CT+
Sbjct: 180 KKKNKFKRKKPDSNSKPRT-GKRPL---RVPGMKPGDS---CFICKGLDHIAKFCTQKAE 338
Query: 272 ---EKKCFRCGQKGHLLADCKRGDV-----VCYNCNEEGHISTQCTQPKKDRTGGKVFA 322
K C RC ++GH +C G YNC + GH C P ++ GG +FA
Sbjct: 339 WEKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQE--GGTMFA 509
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 45.4 bits (106), Expect = 2e-04
Identities = 31/113 (27%), Positives = 52/113 (45%), Gaps = 4/113 (3%)
Frame = +2
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDG +ER Q ++ +LR V W LP ++ N ++SSIG PF +G
Sbjct: 677 QTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHV 856
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVK----MIQEKMKVSHSRQKSYHDKRR 1450
++G V G Q +++K IQ ++ + R ++ +KRR
Sbjct: 857 DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRR 1015
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 45.1 bits (105), Expect = 2e-04
Identities = 22/57 (38%), Positives = 29/57 (50%)
Frame = +2
Query: 1343 TDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQ 1399
+DGQSE + E LR + W P E+ YN Y+ S M PF+ALYG+
Sbjct: 104 SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWYNTSYNISAAMTPFKALYGR 274
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 44.7 bits (104), Expect = 3e-04
Identities = 23/78 (29%), Positives = 37/78 (46%), Gaps = 2/78 (2%)
Frame = +2
Query: 43 PAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPV--LEQ 100
P F G PD WL+ +ER+F+ + +E +KV+ L + A WW +L E
Sbjct: 347 PDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEG 526
Query: 101 DEAVVTWAVFRREFLNRY 118
+ TW R++ +Y
Sbjct: 527 KSKIKTWDKMRQKLTRKY 580
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 43.1 bits (100), Expect = 8e-04
Identities = 22/88 (25%), Positives = 30/88 (34%), Gaps = 31/88 (35%)
Frame = +3
Query: 255 CFKCGEKGHKSNVCTKDEKK-----------------CFRCGQKGHLLADCKRGD----- 292
C+ CG+ GH + C + ++ C+ CG H DC RG
Sbjct: 351 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 530
Query: 293 ---------VVCYNCNEEGHISTQCTQP 311
CY C GHI+ C P
Sbjct: 531 GGGGYGGGGTSCYRCGGVGHIARDCATP 614
Score = 32.0 bits (71), Expect = 1.8
Identities = 15/61 (24%), Positives = 20/61 (32%), Gaps = 17/61 (27%)
Frame = +3
Query: 275 CFRCGQKGHLLADCKRG-----------------DVVCYNCNEEGHISTQCTQPKKDRTG 317
C+ CG GH+ DC R D CY C H + C + +
Sbjct: 351 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 530
Query: 318 G 318
G
Sbjct: 531 G 533
Score = 30.8 bits (68), Expect = 4.0
Identities = 14/44 (31%), Positives = 22/44 (49%), Gaps = 4/44 (9%)
Frame = +3
Query: 279 GQKGHLLADCKRGDVVCYNCNEEGHISTQCTQ----PKKDRTGG 318
G +G + + G CY C + GHI+ C + + DR+GG
Sbjct: 303 GGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG 434
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 42.4 bits (98), Expect = 0.001
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 2/78 (2%)
Frame = -2
Query: 43 PAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWW--VSLLPVLEQ 100
P F+G PD WL+ +ER+F+ + E +KV+ L + A WW V E
Sbjct: 724 PDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREG 545
Query: 101 DEAVVTWAVFRREFLNRY 118
+ TW R++ +Y
Sbjct: 544 KSKIKTWEKMRQKLTRKY 491
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 39.7 bits (91), Expect = 0.009
Identities = 21/78 (26%), Positives = 37/78 (46%), Gaps = 2/78 (2%)
Frame = +2
Query: 43 PAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQD- 101
P F+G D WL+ +ER+F + E +KV+ L + A WW +L +++
Sbjct: 629 PDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREG 808
Query: 102 -EAVVTWAVFRREFLNRY 118
+ TW R++ +Y
Sbjct: 809 KSKIKTWDKMRQKLTRKY 862
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 38.1 bits (87), Expect = 0.025
Identities = 38/104 (36%), Positives = 48/104 (45%)
Frame = +3
Query: 775 FIMMRLSWAWVVF*CKRAKW*LMLLGS*GFMKRIILHMIWSWLQWCLF*KFGGIICMVSD 834
FI RLS V +* + GS* M+ MI WL+ *+FG CMV
Sbjct: 54 FIQTRLSLD*VAY*PSMRRSSPTRQGS*ENMRETTPPMI*KWLR*YSP*RFGAHTCMVPR 233
Query: 835 LKYSAITGV*SIYLTRRN*T*DRGDGLSC*KIMILV*ITILVKL 878
+Y V*SI+L +*T* RG G + * I + I KL
Sbjct: 234 FRYIRTIKV*SIFLPSLS*T*GRGGGWNS*LTTI*TSLIIREKL 365
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 35.8 bits (81), Expect = 0.12
Identities = 16/57 (28%), Positives = 20/57 (35%), Gaps = 20/57 (35%)
Frame = +1
Query: 255 CFKCGEKGHKSNVCTKDEK--------------------KCFRCGQKGHLLADCKRG 291
C+ CGE GH + CT C+ CG+ GH DC G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 33.5 bits (75), Expect = 0.62
Identities = 14/54 (25%), Positives = 20/54 (36%), Gaps = 20/54 (37%)
Frame = +1
Query: 275 CFRCGQKGHLLADCKRGDV--------------------VCYNCNEEGHISTQC 308
C+ CG+ GH+ +C G CY+C E GH + C
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
Score = 30.4 bits (67), Expect = 5.2
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +1
Query: 295 CYNCNEEGHISTQCT 309
CYNC E GH++ +CT
Sbjct: 16 CYNCGESGHMARECT 60
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 35.4 bits (80), Expect = 0.16
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 6/65 (9%)
Frame = +1
Query: 453 PDEIPDVPPEREV-EFSIDLVP----GTKPMSMAP-YRMSSSELAELKKQLED*LDKKFV 506
P++ VP R + + +I L+P P+ P Y MS EL LKK LED LDK F+
Sbjct: 373 PEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKTLEDLLDKGFI 552
Query: 507 RPNVS 511
+ + S
Sbjct: 553 KASGS 567
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 33.9 bits (76), Expect = 0.47
Identities = 14/48 (29%), Positives = 20/48 (41%), Gaps = 13/48 (27%)
Frame = -1
Query: 274 KCFRCGQKGHLLADCKRGDVV-------------CYNCNEEGHISTQC 308
KC++C Q GH ++C CY CN+ GH + C
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNC 361
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 33.9 bits (76), Expect = 0.47
Identities = 23/64 (35%), Positives = 28/64 (42%), Gaps = 8/64 (12%)
Frame = +1
Query: 213 SERRG----KGRQSRPKPYSAPADK----GKQRLNDERRPNRRDASAEIVCFKCGEKGHK 264
S RRG K S KP + D G+Q PNR A CF CGE GH+
Sbjct: 25 SSRRGGRSYKSGNSWSKPERSSRDDWLIGGRQSSRSSSSPNRSFAGT---CFTCGESGHR 195
Query: 265 SNVC 268
++ C
Sbjct: 196 ASDC 207
Score = 30.4 bits (67), Expect = 5.2
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 2/20 (10%)
Frame = +1
Query: 275 CFRCGQKGHLLADC--KRGD 292
CF CG+ GH +DC KRGD
Sbjct: 166 CFTCGESGHRASDCPNKRGD 225
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 32.3 bits (72), Expect = 1.4
Identities = 14/29 (48%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Frame = -1
Query: 295 CYNCNEEGHISTQCTQ---PKKDRTGGKV 320
CY+C+E GHI+ CT KK + G KV
Sbjct: 316 CYSCHERGHIARNCTNTSAAKKKKKGPKV 230
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.351 0.154 0.543
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,455,465
Number of Sequences: 36976
Number of extensions: 773255
Number of successful extensions: 6387
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2522
Number of HSP's successfully gapped in prelim test: 290
Number of HSP's that attempted gapping in prelim test: 3582
Number of HSP's gapped (non-prelim): 3188
length of query: 1454
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1346
effective length of database: 5,021,319
effective search space: 6758695374
effective search space used: 6758695374
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC130807.5


