

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.11 - phase: 0
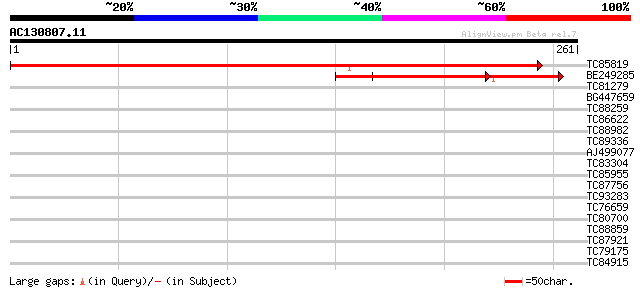
(261 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85819 similar to GP|13194678|gb|AAK15504.1 unknown {Pennisetum... 466 e-132
BE249285 similar to GP|9293961|dbj| gene_id:MAL21.7~unknown prot... 144 3e-35
TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.... 35 0.029
BG447659 weakly similar to GP|9759160|dbj contains similarity to... 33 0.14
TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Ar... 32 0.25
TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myo... 32 0.32
TC88982 weakly similar to GP|15450828|gb|AAK96685.1 putative AAA... 32 0.32
TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Ara... 31 0.42
AJ499077 similar to GP|21684652|gb putative glutamate receptor p... 29 1.6
TC83304 homologue to GP|19386835|dbj|BAB86213. putative deoxygua... 29 2.1
TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {... 29 2.1
TC87756 similar to GP|9665146|gb|AAF97330.1| Unknown protein {Ar... 29 2.1
TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein... 29 2.1
TC76659 weakly similar to PIR|T09127|T09127 probable erythrocyte... 29 2.1
TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity... 28 3.6
TC88859 similar to GP|9758397|dbj|BAB08884.1 signal recognition ... 28 4.6
TC87921 similar to SP|Q9ZQX4|VATF_ARATH Probable vacuolar ATP sy... 27 6.1
TC79175 similar to GP|21592883|gb|AAM64833.1 cytochrome-b5 reduc... 27 7.9
TC84915 similar to GP|15724222|gb|AAL06504.1 At1g28510/F3M18_5 {... 27 7.9
>TC85819 similar to GP|13194678|gb|AAK15504.1 unknown {Pennisetum ciliare},
partial (15%)
Length = 1597
Score = 466 bits (1200), Expect = e-132
Identities = 244/278 (87%), Positives = 244/278 (87%), Gaps = 33/278 (11%)
Frame = +3
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKF SAAEDVIDHLRTENDKLLGQIN
Sbjct: 762 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFDSAAEDVIDHLRTENDKLLGQIN 941
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN 120
DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN
Sbjct: 942 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN 1121
Query: 121 NTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEG------------------------- 155
NTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEG
Sbjct: 1122 NTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGNSVDRESTRSFLKENASNKRQECSS 1301
Query: 156 --------VDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWI 207
VDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWI
Sbjct: 1302 SKANDQSGVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWI 1481
Query: 208 SRAPGEEAELLYHVLSLGTLERLAPEWMREDIMFSPTM 245
SRAPGEEAELLYHVLSLGTLERLAPEWMREDIMFSPTM
Sbjct: 1482 SRAPGEEAELLYHVLSLGTLERLAPEWMREDIMFSPTM 1595
>BE249285 similar to GP|9293961|dbj| gene_id:MAL21.7~unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 454
Score = 144 bits (363), Expect = 3e-35
Identities = 67/71 (94%), Positives = 69/71 (96%)
Frame = +3
Query: 151 SENEGVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRA 210
++ GVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRA
Sbjct: 138 NDQSGVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRA 317
Query: 211 PGEEAELLYHV 221
PGEEAELLYHV
Sbjct: 318 PGEEAELLYHV 350
Score = 74.7 bits (182), Expect = 3e-14
Identities = 45/89 (50%), Positives = 54/89 (60%), Gaps = 1/89 (1%)
Frame = +1
Query: 168 VHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLYHV-LSLGT 226
+H + Y L +KL D C++ + S Y P E + Y + +SLGT
Sbjct: 208 IHWI*NYPLTVKL--DDYVSLQCINQVVTPSVYHGLAE-----PLERKQNCYTMFVSLGT 366
Query: 227 LERLAPEWMREDIMFSPTMCPIFFERVTH 255
LERLAPEWMREDIMFSPTMCPIFFERVTH
Sbjct: 367 LERLAPEWMREDIMFSPTMCPIFFERVTH 453
>TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.170 -
Arabidopsis thaliana, partial (31%)
Length = 1205
Score = 35.0 bits (79), Expect = 0.029
Identities = 33/146 (22%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Frame = +2
Query: 29 EQELKFLKFV-SAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAK-DNELVEHKRLLLE 86
++ELK LK + L E ++ + +I L ELT R + N + KRL+
Sbjct: 95 QEELKLLKLERDKTTTEVRQLHKELNEKVSEIKRLQLELTRQRSKEASNAMDSSKRLIET 274
Query: 87 EKKKNEALFEEVEKLQKLLKEGTSGDLS----------NRKVVNNTSNNSSIRMTRKRMR 136
+K+N L E +L+ +K ++ DLS + ++ +N + ++R +
Sbjct: 275 LEKENTTLKMEKSELEAAVKASSASDLSDPSKSFPGKEDMEISLQKMSNDLKKTQQERDK 454
Query: 137 QEQDALDIEARCIPSENEGVDTQESD 162
Q+ ++ + ENE + + D
Sbjct: 455 AVQELTRLKQHLLEKENEESEKMDED 532
>BG447659 weakly similar to GP|9759160|dbj contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (11%)
Length = 665
Score = 32.7 bits (73), Expect = 0.14
Identities = 40/185 (21%), Positives = 83/185 (44%), Gaps = 1/185 (0%)
Frame = +1
Query: 3 ALYKKLYSKYTTLKTNKL-SELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIND 61
AL K+ + + L+ +L +EL + + ++ ++ + + ++ + D L+ +I
Sbjct: 124 ALSKESSQEASHLEIERLKAELAALARHVDVSDMELQTLRKQIVKESKRGQD-LMKEIII 300
Query: 62 LGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNN 121
L +E +++ DN HKR+ + K +N + E + E +L+ K
Sbjct: 301 LKDERDALKTECDNVRSFHKRMD-DAKVRNRSQLESGD--HHAFVEEIRQELNYEK---- 459
Query: 122 TSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLS 181
N+++R+ K+M++ L + + + E + S H N H L+MKLS
Sbjct: 460 -DTNANLRLQLKKMQESNAELVLAVQDLEEMLEQKNMNMSKHSNGQEHNKNSQELEMKLS 636
Query: 182 TDCQT 186
C+T
Sbjct: 637 -QCET 648
>TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1757
Score = 32.0 bits (71), Expect = 0.25
Identities = 30/120 (25%), Positives = 54/120 (45%), Gaps = 5/120 (4%)
Frame = +3
Query: 53 DKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE----- 107
+ ++ Q + + E S +NE+ K+ L ++KN L + V L LKE
Sbjct: 480 EDMVKQHSKVAEEAISGWEKAENEVSSLKQQLDAARQKNSGLEDRVSHLDGALKECMRQL 659
Query: 108 GTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWL 167
+ ++ +K+ +NNS +R R E+ ++EA+ S+ E + SD Q L
Sbjct: 660 RQAREVQEQKIHEAVANNSHDSGSR-RFELERKVAELEAQLQTSKAEAAASIRSDLQRRL 836
>TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myosin heavy
chain {Arabidopsis thaliana}, partial (17%)
Length = 2546
Score = 31.6 bits (70), Expect = 0.32
Identities = 53/203 (26%), Positives = 85/203 (41%), Gaps = 21/203 (10%)
Frame = +2
Query: 20 LSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLG-------QINDLGNELTSVRVA 72
LS+L + E E + + AE +EN+ L+G +I++L L SV
Sbjct: 614 LSKLASLQSENE-DLKRQIVEAEKKTSQSFSENELLVGTNIQLKTKIDELQESLNSVVSE 790
Query: 73 KD---NELVEHKRLLLEEK----------KKNEALFEEVE-KLQKLLKEGTSGDLSNRKV 118
K+ ELV HK LL E NE EVE KLQ+ L++ T + S K
Sbjct: 791 KEVTAQELVSHKNLLAELNDVQSKSSEIHSANEVRILEVESKLQEALQKHTEKE-SETKE 967
Query: 119 VNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDM 178
+N N ++ ++ +EQ + A +EN + +ES + + A E +
Sbjct: 968 LNEKLNTLEGQI---KIYEEQAHEAVAA----AENRKAELEESLIKLKHLEAAVEEQQNK 1126
Query: 179 KLSTDCQTGRLCLSAMHQSSGYS 201
L + +T + + SG S
Sbjct: 1127SLERETETAGINEEKLKLCSGNS 1195
Score = 28.1 bits (61), Expect = 3.6
Identities = 30/100 (30%), Positives = 52/100 (52%), Gaps = 15/100 (15%)
Frame = +3
Query: 18 NKLSELED------VHKEQELK-FLKFVSAAEDVIDHLRTENDKLLGQIN------DLGN 64
+KLS+L+ V K++ +K L +AAED++ E L QI+ +L N
Sbjct: 1206 SKLSDLQSKLSAALVEKDETVKEILASKNAAEDLVTQHNEEVQTLKSQISSVIDDRNLLN 1385
Query: 65 ELTSVRVAKDNE--LVEHKRLLLEEKKKNEALFEEVEKLQ 102
E T+ + K+ E +++ + L E +K ++L EVE L+
Sbjct: 1386 E-TNQNLKKELESIILDLEEKLKEHQKNEDSLKSEVETLK 1502
>TC88982 weakly similar to GP|15450828|gb|AAK96685.1 putative AAA-type
ATPase {Arabidopsis thaliana}, partial (34%)
Length = 859
Score = 31.6 bits (70), Expect = 0.32
Identities = 33/164 (20%), Positives = 69/164 (41%), Gaps = 13/164 (7%)
Frame = +2
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQEL-----KFLKFVSAAEDVIDHLRTENDKLLGQIN 60
KK++ + ++L+EL+ EL + AE+ + ++ +N + Q+
Sbjct: 347 KKVFDLMRKQEQSRLAELDAEKVNYELIQTQGDIDRLRKMAEEQRNLIQEQNQRQ-AQVL 523
Query: 61 DLGNELTSVRVAKDNE--------LVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGD 112
+EL R+ D+E LV+ + K++ EE + QKLL E +
Sbjct: 524 RFEDELARKRMQTDHEDQRRHNVELVQMQEKSFVRKEQARKDSEEQMQAQKLLTEQKKAE 703
Query: 113 LSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGV 156
+ + N+ R+ K + +EQ+ +++ C + G+
Sbjct: 704 IDKETIRAKEKANAEKRIRLKVLTEEQNRRELKTNCRGKQINGL 835
>TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 1241
Score = 31.2 bits (69), Expect = 0.42
Identities = 33/122 (27%), Positives = 56/122 (45%), Gaps = 3/122 (2%)
Frame = +1
Query: 26 VHKEQELKF--LKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRL 83
+ E+EL+ LK + E++I L+ +N+K GQ++ + ELT +EL K
Sbjct: 487 IKNEEELRVSNLKLKLSEEEIIK-LKNQNEKSEGQLDSVQKELT----LNMDELEHKKGQ 651
Query: 84 LLEEKKKNEALFEEVEKLQKLLKEGTSG-DLSNRKVVNNTSNNSSIRMTRKRMRQEQDAL 142
+LE +K+ L V L + L+ +SN +V + RQ QD L
Sbjct: 652 VLELQKQKAELETHVPNLVEQLEVANEHLKISNDEVAR---LRKELESNLAETRQLQDQL 822
Query: 143 DI 144
++
Sbjct: 823 EV 828
>AJ499077 similar to GP|21684652|gb putative glutamate receptor protein
GLR3.4b {Arabidopsis thaliana}, partial (15%)
Length = 508
Score = 29.3 bits (64), Expect = 1.6
Identities = 16/47 (34%), Positives = 23/47 (48%)
Frame = -3
Query: 87 EKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRK 133
E K E + +L + + RK++NN+ N SSIR TRK
Sbjct: 293 ESKSPTTFSEHISNGSELAVGTRHKEFNIRKLINNSHNPSSIRSTRK 153
>TC83304 homologue to GP|19386835|dbj|BAB86213. putative deoxyguanosine
kinase {Oryza sativa (japonica cultivar-group)}, partial
(15%)
Length = 919
Score = 28.9 bits (63), Expect = 2.1
Identities = 16/31 (51%), Positives = 19/31 (60%), Gaps = 1/31 (3%)
Frame = -3
Query: 74 DNELVEHK-RLLLEEKKKNEALFEEVEKLQK 103
D + EHK R EEKKK +FEE +K QK
Sbjct: 347 DKNMYEHKKRWNNEEKKKKRGIFEEGKKPQK 255
>TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {Cicer
arietinum}, partial (98%)
Length = 1981
Score = 28.9 bits (63), Expect = 2.1
Identities = 13/39 (33%), Positives = 24/39 (61%)
Frame = +2
Query: 79 EHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRK 117
+HK ++KK+ L +EVE K++++G + D S +K
Sbjct: 1511 DHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKK 1627
>TC87756 similar to GP|9665146|gb|AAF97330.1| Unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 2783
Score = 28.9 bits (63), Expect = 2.1
Identities = 35/161 (21%), Positives = 65/161 (39%), Gaps = 6/161 (3%)
Frame = +1
Query: 9 YSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTEN------DKLLGQINDL 62
Y + T +S+L+ + E E+ VS DV H + D + N++
Sbjct: 565 YKEVALAPTGTISKLQVYNPENEIS----VSQEHDVGKHEEEDIEAHRNIDPTPKEANNV 732
Query: 63 GNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNT 122
E + ++ E + ++ EKK+ L + V+ + SGD+ + V+N+
Sbjct: 733 FKEKSDDSLSDSIEDSQDDTVVSTEKKEETQLNKVVQDSCATAEGLESGDVEAQGAVDNS 912
Query: 123 SNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDH 163
++ + +QE A D+ PS+N T S H
Sbjct: 913 IVIDAVEDAMESYKQELVASDLPCSFEPSDN----TSSSPH 1023
>TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein
{Arabidopsis thaliana}, partial (18%)
Length = 902
Score = 28.9 bits (63), Expect = 2.1
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 3/110 (2%)
Frame = +1
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAE---DVIDHLRTENDKLLGQINDL 62
K L+SK K L ++++ + ++LK AE ++IDHL KLL +N
Sbjct: 580 KPLFSKALAKKEKSLEDVQN--QIEQLKGSIATKKAEMGTELIDHLTPXEKKLLSDLNPE 753
Query: 63 GNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGD 112
+L VA + +E + E + L + K ++ L+ G S D
Sbjct: 754 IKDLKEKLVACKTDRIESEARKAELETN---LTTNLXKXKQELEAGISSD 894
>TC76659 weakly similar to PIR|T09127|T09127 probable erythrocyte-binding
protein MAEBL - Plasmodium yoelii, partial (2%)
Length = 843
Score = 28.9 bits (63), Expect = 2.1
Identities = 20/69 (28%), Positives = 35/69 (49%)
Frame = +1
Query: 79 EHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQE 138
E K+ L EEK+ E L +E+E+ +K +E RK V + RK++++
Sbjct: 553 EDKKFLEEEKEAEEELLKEIEEDRKQAEE------EARKKVKRARKAEEV--ARKKVKRA 708
Query: 139 QDALDIEAR 147
+ A + EA+
Sbjct: 709 KKAEEAEAK 735
>TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (4%)
Length = 958
Score = 28.1 bits (61), Expect = 3.6
Identities = 27/99 (27%), Positives = 44/99 (44%), Gaps = 1/99 (1%)
Frame = +2
Query: 49 RTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEAL-FEEVEKLQKLLKE 107
R+E + L Q NDL + L K+N + L E KKK++AL + E + + K K
Sbjct: 2 RSELEALKAQYNDLKHSLIVDETEKENLRQQIFHLNDELKKKDDALTYIETKTIPKNQKS 181
Query: 108 GTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEA 146
+ S T I+M ++ ++ AL+ A
Sbjct: 182 ASIPHNSKEM----TDLREKIKMLEDLIKSKETALEASA 286
>TC88859 similar to GP|9758397|dbj|BAB08884.1 signal recognition particle
68kD protein-like {Arabidopsis thaliana}, partial (23%)
Length = 893
Score = 27.7 bits (60), Expect = 4.6
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 9/78 (11%)
Frame = +1
Query: 39 SAAEDVIDHLRTENDKLLGQINDLGNELTSV-RVAKDNELVEHKRLLLEEKKKNEALFEE 97
S AED + L+ KL G + EL + + N +EH ++EEK+ E + E
Sbjct: 139 SLAEDALGKLQ----KLEGNSKTMVKELEVLCNECRSNSCIEHALGIMEEKRTQENISEG 306
Query: 98 V--------EKLQKLLKE 107
+ E+L+K L E
Sbjct: 307 ISNISLTGAERLEKFLLE 360
>TC87921 similar to SP|Q9ZQX4|VATF_ARATH Probable vacuolar ATP synthase
subunit F (EC 3.6.3.14) (V-ATPase F subunit), partial
(97%)
Length = 758
Score = 27.3 bits (59), Expect = 6.1
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +3
Query: 183 DCQTGRLCLSAMHQSSGY 200
DCQT R C+ +H + GY
Sbjct: 258 DCQTNRGCIQRIHNTGGY 311
>TC79175 similar to GP|21592883|gb|AAM64833.1 cytochrome-b5 reductase-like
protein {Arabidopsis thaliana}, partial (84%)
Length = 1251
Score = 26.9 bits (58), Expect = 7.9
Identities = 15/55 (27%), Positives = 26/55 (47%)
Frame = +2
Query: 113 LSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWL 167
L + K++ N S +R+ +++ +D L +RCIP E Q + WL
Sbjct: 368 LHSDKMLKKNQNMSFVRILLYQIQNPRDILIC*SRCIPKEK*ASILQA*NQVTWL 532
>TC84915 similar to GP|15724222|gb|AAL06504.1 At1g28510/F3M18_5 {Arabidopsis
thaliana}, partial (65%)
Length = 604
Score = 26.9 bits (58), Expect = 7.9
Identities = 33/114 (28%), Positives = 54/114 (46%)
Frame = +1
Query: 36 KFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALF 95
K V AA D+I L + + I ++ S A+ E+ KR L ++KN+ L
Sbjct: 247 KAVQAAVDLIGELFVFSVAVAVLIFEVQRSARSE--ARKEEI--RKRELEAVRQKNDGLA 414
Query: 96 EEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCI 149
EEVE L+ L+E L+ + + N + + RK M++ + LD+ R I
Sbjct: 415 EEVELLKLRLQE--LEQLARGRGLTGILNLQTHYLVRK-MKRRRKLLDLMKRII 567
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,221,418
Number of Sequences: 36976
Number of extensions: 93351
Number of successful extensions: 374
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 370
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 373
length of query: 261
length of database: 9,014,727
effective HSP length: 94
effective length of query: 167
effective length of database: 5,538,983
effective search space: 925010161
effective search space used: 925010161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC130807.11


