

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.14 - phase: 0
(394 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
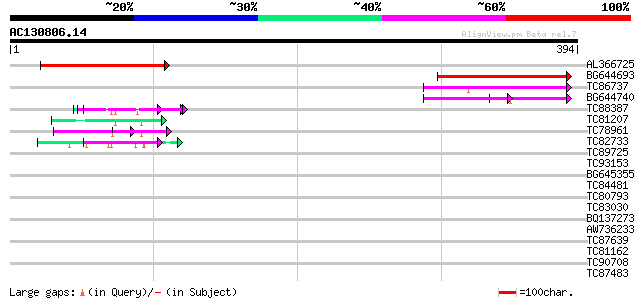
Score E
Sequences producing significant alignments: (bits) Value
AL366725 155 3e-38
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 90 1e-18
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 75 3e-14
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 51 9e-07
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 50 2e-06
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 46 3e-05
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 46 3e-05
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 45 5e-05
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 35 0.038
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 33 0.25
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 32 0.43
TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis t... 31 0.95
TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein cons... 30 1.6
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 30 1.6
BQ137273 30 1.6
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 30 1.6
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 29 2.8
TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein... 29 2.8
TC90708 weakly similar to PIR|S26703|S26703 dnaJ protein homolog... 29 3.6
TC87483 similar to GP|21955291|gb|AAK59492.2 unknown protein {Ar... 29 3.6
>AL366725
Length = 485
Score = 155 bits (391), Expect = 3e-38
Identities = 67/90 (74%), Positives = 76/90 (84%)
Frame = +2
Query: 22 PKPYSAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGH 81
PKPYSAPA KGKQR+ D+RRPK +D PAEIVCF GEKGHKSNVC K+ KKC RC +KGH
Sbjct: 215 PKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEIKKCVRCDKKGH 394
Query: 82 VLADCKRGDIVCYNCNGEGHISSQCTQPKK 111
++ADCKR DIVC+NCN EGHI SQC QPK+
Sbjct: 395 IVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 90.1 bits (222), Expect = 1e-18
Identities = 44/93 (47%), Positives = 63/93 (67%)
Frame = +2
Query: 298 DEIPDVPPEREVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPW 357
D + VPPE +++F IDL+P P+ + YR++ +L LK QL+DLL+K F++ S++P
Sbjct: 2 DHLL*VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 358 GAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
G VL KKKDG +R+ I YP LN V IK +YP
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYP 280
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 75.5 bits (184), Expect = 3e-14
Identities = 47/110 (42%), Positives = 63/110 (56%), Gaps = 7/110 (6%)
Frame = +1
Query: 288 VVNEFPEVFSDEIP-DVPPEREV-EFSIDLVP----RTKPVSMAP-YRMSASELAELKNQ 340
V+ EFP++F+ E VP R + + +I L+P P+ P Y MS EL LK
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 341 LEDLLDKKFVRASVFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
LEDLLDK F++AS GAPVL +K G +R C+ Y LN + K+RYP
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYP 672
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 50.8 bits (120), Expect = 9e-07
Identities = 27/64 (42%), Positives = 37/64 (57%), Gaps = 7/64 (10%)
Frame = -1
Query: 334 LAELKNQLEDLLD-------KKFVRASVFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIK 386
LA+ N++E D K+F + S+ P GA +L +KKDG R+CI Y NKV K
Sbjct: 334 LADGSNRVEKTKDLT*RFARKRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTK 155
Query: 387 NRYP 390
N+YP
Sbjct: 154 NKYP 143
Score = 46.2 bits (108), Expect = 2e-05
Identities = 27/62 (43%), Positives = 37/62 (59%)
Frame = -2
Query: 288 VVNEFPEVFSDEIPDVPPEREVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDK 347
V+ F VF D P +P ERE+ F IDL+ T+ +S P M +EL +LK L+D L+K
Sbjct: 450 VLKGFS*VFPDNFPVIPLEREIFFCIDLLLDTQLISNPP*LMDRTELKKLKI*LKDSLEK 271
Query: 348 KF 349
F
Sbjct: 270 GF 265
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 49.7 bits (117), Expect = 2e-06
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Frame = +1
Query: 48 PAEIVCFKCGEKGHKSNVCTKDEKK------CFRCGQKGHVLADCKRGDIVCYNCNGEGH 101
P E +C CG+ GH++ CT +K C C ++GH+ +C + C NC GH
Sbjct: 580 PNEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC-TNEKACNNCRKTGH 756
Query: 102 ISSQC 106
++ C
Sbjct: 757 LARDC 771
Score = 49.7 bits (117), Expect = 2e-06
Identities = 25/72 (34%), Positives = 38/72 (52%)
Frame = +1
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCTQPKK 111
+C C ++GH + CT +EK C C + GH+ DC D +C CN GH++ QC +
Sbjct: 670 LCNNCYKQGHIAVECT-NEKACNNCRKTGHLARDCPN-DPICNLCNISGHVARQCPKSNV 843
Query: 112 VRTGGKVFALAG 123
+ G +L G
Sbjct: 844 IGDRGGGGSLRG 879
Score = 49.3 bits (116), Expect = 3e-06
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Frame = +1
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCK-----RGDI-VCYNCNGEGHISSQ 105
+C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC +GHI+ +
Sbjct: 535 LCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVE 711
Query: 106 CTQPKKVRTGGKVFALA 122
CT K K LA
Sbjct: 712 CTNEKACNNCRKTGHLA 762
Score = 43.9 bits (102), Expect = 1e-04
Identities = 24/85 (28%), Positives = 33/85 (38%), Gaps = 23/85 (27%)
Frame = +1
Query: 45 RDTPAEIVCFKCGEKGHKSNVCTKD----------------------EKKCFRCGQKGHV 82
RD P + +C C GH + C K + C C Q GH+
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 83 LADCKRGDI-VCYNCNGEGHISSQC 106
DC G + +C NC G GH + +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 35.8 bits (81), Expect = 0.029
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +1
Query: 73 CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCT 107
C C + GH + +C VC+NC+ GHI+S+C+
Sbjct: 424 CKNCKRPGHYVRECPNV-AVCHNCSLPGHIASECS 525
Score = 32.0 bits (71), Expect = 0.43
Identities = 15/44 (34%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Frame = +1
Query: 50 EIVCFKCGEKGHKSNVCTKDEKK-CFRCGQKGHVLADCKRGDIV 92
++VC C + GH S C C CG +GH +C G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 45.8 bits (107), Expect = 3e-05
Identities = 29/111 (26%), Positives = 38/111 (34%), Gaps = 31/111 (27%)
Frame = +3
Query: 30 GKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKK----------------- 72
G G+ ERR G C+ CG+ GH + C + ++
Sbjct: 300 GGGRGFRGGERRNGGGG------CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRA 461
Query: 73 CFRCGQKGHVLADCKRGD--------------IVCYNCNGEGHISSQCTQP 109
C+ CG H DC RG CY C G GHI+ C P
Sbjct: 462 CYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATP 614
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 45.8 bits (107), Expect = 3e-05
Identities = 22/60 (36%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Frame = +2
Query: 31 KGKQRLNDERRPKGRDTP-AEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHVLADCK 87
+G + D R GR P CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 290 RGSRDSRDSREYLGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 469
Score = 45.4 bits (106), Expect = 4e-05
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Frame = +2
Query: 72 KCFRCGQKGHVLADCKRGD--IVCYNCNGEGHISSQC-TQPKKV 112
+CF CG GH DCK GD CY C GHI C PKK+
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKL 487
Score = 28.9 bits (63), Expect = 3.6
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKKCFR 75
C++CGE+GH C KK R
Sbjct: 425 CYRCGERGHIEKNCKNSPKKLSR 493
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 45.1 bits (105), Expect = 5e-05
Identities = 33/123 (26%), Positives = 50/123 (39%), Gaps = 22/123 (17%)
Frame = +3
Query: 20 VGPKPYSAPAGKGKQRLNDER-----RPKGRDTPAEIV-------CFKCGEKGHKSNVCT 67
V P P + P+ K K + ++ +P+ P + CF C H + CT
Sbjct: 147 VDPTPPNDPSKKKKNKFKRKKPDSNSKPRTGKRPLRVPGMKPGDSCFICKGLDHIAKFCT 326
Query: 68 KD-----EKKCFRCGQKGHVLADCKRGDI-----VCYNCNGEGHISSQCTQPKKVRTGGK 117
+ K C RC ++GH +C G YNC GH + C P ++ GG
Sbjct: 327 QKAEWEKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHP--LQEGGT 500
Query: 118 VFA 120
+FA
Sbjct: 501 MFA 509
Score = 44.7 bits (104), Expect = 6e-05
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 12/67 (17%)
Frame = +3
Query: 52 VCFKCGEKGHKSNVCT-----KDEKKCFRCGQKGHVLADC----KRGDIV---CYNCNGE 99
+C +C +GH++ C +D K + CG GH LA+C + G + C+ C +
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 100 GHISSQC 106
GH+S C
Sbjct: 534 GHLSKNC 554
Score = 28.1 bits (61), Expect = 6.1
Identities = 15/60 (25%), Positives = 24/60 (40%), Gaps = 7/60 (11%)
Frame = +3
Query: 54 FKCGEKGHKSNVCTKDEKK-------CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQC 106
+ CG+ GH C ++ CF C ++GH+ +C + Y G I C
Sbjct: 435 YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNCPKNAHGIYPKGGCCKICGXC 614
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 35.4 bits (80), Expect = 0.038
Identities = 16/57 (28%), Positives = 20/57 (35%), Gaps = 20/57 (35%)
Frame = +1
Query: 53 CFKCGEKGHKSNVCTK--------------------DEKKCFRCGQKGHVLADCKRG 89
C+ CGE GH + CT C+ CG+ GH DC G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 27.7 bits (60), Expect = 8.0
Identities = 8/15 (53%), Positives = 11/15 (73%)
Frame = +1
Query: 93 CYNCNGEGHISSQCT 107
CYNC GH++ +CT
Sbjct: 16 CYNCGESGHMARECT 60
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 32.7 bits (73), Expect = 0.25
Identities = 23/53 (43%), Positives = 26/53 (48%), Gaps = 5/53 (9%)
Frame = +2
Query: 1 MAGRNDAAIAAALEAVAQAVGPKPYSAPAGKGKQRL-----NDERRPKGRDTP 48
M G +DAA+ AALEAVAQAV P G + L N KGR P
Sbjct: 2 MTGSSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAP 160
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 32.0 bits (71), Expect = 0.43
Identities = 14/48 (29%), Positives = 20/48 (41%), Gaps = 13/48 (27%)
Frame = -1
Query: 72 KCFRCGQKGHVLADCKRGDIV-------------CYNCNGEGHISSQC 106
KC++C Q GH ++C CY CN GH ++ C
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNC 361
>TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis thaliana},
partial (23%)
Length = 727
Score = 30.8 bits (68), Expect = 0.95
Identities = 9/18 (50%), Positives = 15/18 (83%)
Frame = +2
Query: 70 EKKCFRCGQKGHVLADCK 87
++KC+ CGQ GH+ A+C+
Sbjct: 200 QEKCYMCGQVGHLAAECR 253
>TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein constans-like
14. [Mouse-ear cress] {Arabidopsis thaliana}, partial
(27%)
Length = 937
Score = 30.0 bits (66), Expect = 1.6
Identities = 15/66 (22%), Positives = 28/66 (41%), Gaps = 3/66 (4%)
Frame = +1
Query: 49 AEIVCFKCGEKGHKSNVCTKDEKK---CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQ 105
A +C C H +N + + C RC + ++ + +C NC+ GH +S
Sbjct: 298 AACLCLSCDRNVHSANTLARRHSRTLLCERCSSQPALVRCSEEKVSLCQNCDWLGHGNST 477
Query: 106 CTQPKK 111
+ K+
Sbjct: 478 SSNHKR 495
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 30.0 bits (66), Expect = 1.6
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 2/20 (10%)
Frame = +1
Query: 73 CFRCGQKGHVLADC--KRGD 90
CF CG+ GH +DC KRGD
Sbjct: 166 CFTCGESGHRASDCPNKRGD 225
>BQ137273
Length = 699
Score = 30.0 bits (66), Expect = 1.6
Identities = 14/41 (34%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Frame = +1
Query: 154 HCFIALECAYKLGLIVS-DMKGEMVVETPAKGSVTTSLVCL 193
HC + C++ LG + M+ +++ ETPA+ VT ++CL
Sbjct: 388 HCVPSSMCSFMLGYACNCTMRIKLLQETPAQNCVTYGMLCL 510
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 30.0 bits (66), Expect = 1.6
Identities = 19/62 (30%), Positives = 27/62 (42%)
Frame = +1
Query: 17 AQAVGPKPYSAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRC 76
+Q VG + Y+A G+G+ R R +GR + C C H + C CFR
Sbjct: 196 SQEVGTEYYNAGRGRGRGR--GRGRGRGRSNSNRLQCQICARNNHDAARC------CFRY 351
Query: 77 GQ 78
Q
Sbjct: 352 DQ 357
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 29.3 bits (64), Expect = 2.8
Identities = 13/29 (44%), Positives = 17/29 (57%), Gaps = 3/29 (10%)
Frame = -1
Query: 93 CYNCNGEGHISSQCTQ---PKKVRTGGKV 118
CY+C+ GHI+ CT KK + G KV
Sbjct: 316 CYSCHERGHIARNCTNTSAAKKKKKGPKV 230
>TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein T26I12.220
- Arabidopsis thaliana, partial (9%)
Length = 756
Score = 29.3 bits (64), Expect = 2.8
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +2
Query: 83 LADCKRGDIVCYNCNGEGHISSQCTQPK 110
+A KR + +CY C +GH S+C P+
Sbjct: 392 VASGKRKNRMCYGCRQKGHNLSECPNPQ 475
>TC90708 weakly similar to PIR|S26703|S26703 dnaJ protein homolog YDJ1 -
yeast (Saccharomyces cerevisiae), partial (43%)
Length = 1098
Score = 28.9 bits (63), Expect = 3.6
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 6/58 (10%)
Frame = +2
Query: 51 IVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDI------VCYNCNGEGHI 102
I+C KC +G K K+C C G + G + VC +CNGEG +
Sbjct: 704 IICSKCEGRGGKEGAV----KRCTGCDGHGMKTMMRQMGPMIQRFQTVCPDCNGEGEM 865
>TC87483 similar to GP|21955291|gb|AAK59492.2 unknown protein {Arabidopsis
thaliana}, partial (97%)
Length = 679
Score = 28.9 bits (63), Expect = 3.6
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 5/82 (6%)
Frame = +3
Query: 72 KCFRCGQKGHVLADCKR---GDIVCYNCNGEGHISSQCTQPKKVRTGGKVFALAGMQTTN 128
+C C G V C R GDI C C+G G + C+ TG + A ++ TN
Sbjct: 246 RCPSCNGTGRVPCLCSRWSDGDIGCSTCSGSGRMG--CSSCGGSGTGRPLPARITIRPTN 419
Query: 129 EDRLIRGT--CFFNSIPLIAII 148
+ G+ F+S +A++
Sbjct: 420 RPS*LTGSV*SLFSSQLCLALV 485
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,147,480
Number of Sequences: 36976
Number of extensions: 202063
Number of successful extensions: 1053
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 960
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1024
length of query: 394
length of database: 9,014,727
effective HSP length: 98
effective length of query: 296
effective length of database: 5,391,079
effective search space: 1595759384
effective search space used: 1595759384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC130806.14


