

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC129092.1 - phase: 0
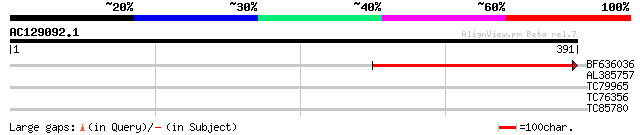
(391 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF636036 similar to GP|6069471|dbj S-locus protein 3 {Brassica r... 288 2e-78
AL385757 weakly similar to GP|17979018|gb| GTP cyclohydrolase I ... 30 1.6
TC79965 similar to GP|7549635|gb|AAF63820.1| hypothetical protei... 28 4.7
TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown prot... 28 6.1
TC85780 similar to SP|P49608|ACOC_CUCMA Aconitate hydratase cyt... 28 7.9
>BF636036 similar to GP|6069471|dbj S-locus protein 3 {Brassica rapa},
partial (50%)
Length = 674
Score = 288 bits (738), Expect = 2e-78
Identities = 139/141 (98%), Positives = 140/141 (98%)
Frame = -2
Query: 251 QSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQKDRKDNTGPLLLD 310
+SLEAFLQ KSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQKDRKDNTGPLLLD
Sbjct: 664 RSLEAFLQXKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQKDRKDNTGPLLLD 485
Query: 311 DSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESNLGWEFQQNNAVDGLY 370
DSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESNLGWEFQQNNAVDGLY
Sbjct: 484 DSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESNLGWEFQQNNAVDGLY 305
Query: 371 FDENDEFAPVVEMLDDEAHAV 391
FDENDEFAPVVEMLDDEAHAV
Sbjct: 304 FDENDEFAPVVEMLDDEAHAV 242
>AL385757 weakly similar to GP|17979018|gb| GTP cyclohydrolase I
{Lycopersicon esculentum}, partial (7%)
Length = 575
Score = 30.0 bits (66), Expect = 1.6
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 8/68 (11%)
Frame = -2
Query: 34 FSVGPTFKGIKMIPPGTHFVYYSSSTRDGKEFSPMIGF---FIDAGP-----SEVIVRKW 85
F +G T+K + + PP H+ YS+ K S +GF F++ G +E+++ KW
Sbjct: 256 FPLGDTYK-M*LXPPNNHYSXYSTLHNRWKSVSSNLGFVRPFLELGSLRAINNELLIVKW 80
Query: 86 DQQEERLV 93
Q + ++
Sbjct: 79 TFQLDSIL 56
>TC79965 similar to GP|7549635|gb|AAF63820.1| hypothetical protein
{Arabidopsis thaliana}, partial (15%)
Length = 742
Score = 28.5 bits (62), Expect = 4.7
Identities = 10/29 (34%), Positives = 16/29 (54%)
Frame = +1
Query: 312 SWFSTDSFLHHHCKEFFSLVLDGSVIDGD 340
SW + LH K+ ++ DGS++D D
Sbjct: 463 SWIMLSNCLHFAVKKIVRIIADGSIVDSD 549
>TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown protein
{Arabidopsis thaliana}, partial (36%)
Length = 2619
Score = 28.1 bits (61), Expect = 6.1
Identities = 17/66 (25%), Positives = 26/66 (38%)
Frame = +3
Query: 62 GKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYNL 121
G F +G+ +DA W + V +E D++Y + + GP NL
Sbjct: 1419 GISFVGDVGYIVDAW--------WHDGWWEGIVVQKESDDKYHVYFPGEKKMSIFGPCNL 1574
Query: 122 SHYEDW 127
H DW
Sbjct: 1575 RHSRDW 1592
>TC85780 similar to SP|P49608|ACOC_CUCMA Aconitate hydratase cytoplasmic (EC
4.2.1.3) (Citrate hydro-lyase) (Aconitase)., partial
(97%)
Length = 3124
Score = 27.7 bits (60), Expect = 7.9
Identities = 16/37 (43%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Frame = +3
Query: 19 LDVPQYTLVAIDTQVFSVG--PTFKGIKMIPPGTHFV 53
L VP L A D+ + P FKG+ M PPG+H V
Sbjct: 1965 LSVPSGNLYAWDSTSTYIHEPPYFKGMSMSPPGSHGV 2075
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,902,864
Number of Sequences: 36976
Number of extensions: 164343
Number of successful extensions: 753
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 749
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 752
length of query: 391
length of database: 9,014,727
effective HSP length: 98
effective length of query: 293
effective length of database: 5,391,079
effective search space: 1579586147
effective search space used: 1579586147
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC129092.1


