

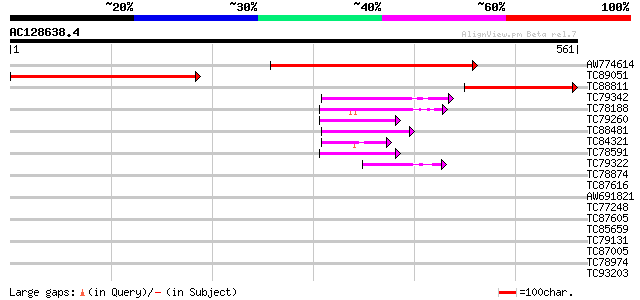
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
(561 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW774614 similar to PIR|T45879|T45 hypothetical protein F4P12.90... 407 e-114
TC89051 similar to GP|4371285|gb|AAD18143.1| unknown protein {Ar... 359 1e-99
TC88811 similar to PIR|T45879|T45879 hypothetical protein F4P12.... 216 1e-56
TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein h... 55 9e-08
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 53 4e-07
TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat pr... 52 6e-07
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 50 2e-06
TC84321 GP|22138476|gb|AAM93460.1 hypothetical protein {Oryza sa... 45 6e-05
TC78591 similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-l... 45 7e-05
TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 42 6e-04
TC78874 similar to PIR|T49098|T49098 hypothetical protein F4F15.... 41 0.001
TC87616 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -... 41 0.001
AW691821 similar to GP|10177570|db contains similarity to regula... 39 0.004
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 39 0.007
TC87605 homologue to PIR|G96563|G96563 probable coatomer complex... 38 0.009
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 38 0.012
TC79131 similar to GP|14031063|gb|AAK52092.1 WD-40 repeat protei... 38 0.012
TC87005 similar to GP|9294061|dbj|BAB02018.1 WD40-repeat protein... 37 0.026
TC78974 similar to GP|22137238|gb|AAM91464.1 AT5g14050/MUA22_5 {... 36 0.034
TC93203 36 0.045
>AW774614 similar to PIR|T45879|T45 hypothetical protein F4P12.90 -
Arabidopsis thaliana, partial (32%)
Length = 615
Score = 407 bits (1047), Expect = e-114
Identities = 194/205 (94%), Positives = 195/205 (94%)
Frame = +1
Query: 259 ADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHICQGSINSISFSAD 318
ADGN YVYEKNKDGAGESSFPILKDQTLFSVAHARY KSNPIARWHICQGSINSISFS D
Sbjct: 1 ADGNTYVYEKNKDGAGESSFPILKDQTLFSVAHARYRKSNPIARWHICQGSINSISFSTD 180
Query: 319 GAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSM 378
GAYLATVGRDGYLRVFDYTKEHLVCGGK YYGGLLCCAWSMDGKYILTGGEDDLVQV SM
Sbjct: 181 GAYLATVGRDGYLRVFDYTKEHLVCGGKRYYGGLLCCAWSMDGKYILTGGEDDLVQVRSM 360
Query: 379 EDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEI 438
E RKVVAWGEGHNSWVSGVA DSYW+SPNSNDNGETITYRFGSVGQDTQLLLWELEMDEI
Sbjct: 361 EHRKVVAWGEGHNSWVSGVALDSYWTSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEI 540
Query: 439 VVPLRRGPPGGSPTFGSPTFSAGSQ 463
VP RRGPPGGS TFGSPTFSAGSQ
Sbjct: 541 AVP*RRGPPGGSATFGSPTFSAGSQ 615
>TC89051 similar to GP|4371285|gb|AAD18143.1| unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 668
Score = 359 bits (922), Expect = 1e-99
Identities = 185/189 (97%), Positives = 185/189 (97%), Gaps = 1/189 (0%)
Frame = +1
Query: 1 MINTTTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNH 60
MINTTTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNH
Sbjct: 100 MINTTTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNH 279
Query: 61 GKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSS 120
GKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSS
Sbjct: 280 GKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSS 459
Query: 121 KSNGGASRIGSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFS 180
KSNGGASRIGSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHF
Sbjct: 460 KSNGGASRIGSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFQ 639
Query: 181 -NSNPVCHA 188
NPVCHA
Sbjct: 640 VIPNPVCHA 666
>TC88811 similar to PIR|T45879|T45879 hypothetical protein F4P12.90 -
Arabidopsis thaliana, partial (14%)
Length = 781
Score = 216 bits (551), Expect = 1e-56
Identities = 108/111 (97%), Positives = 108/111 (97%)
Frame = +3
Query: 451 PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQES 510
P SPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQES
Sbjct: 6 PNI*SPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQES 185
Query: 511 VLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLSSKISNSIYKQ 561
VLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLSSKISNSIYKQ
Sbjct: 186 VLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLSSKISNSIYKQ 338
>TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein homolog
{Arabidopsis thaliana}, partial (47%)
Length = 873
Score = 54.7 bits (130), Expect = 9e-08
Identities = 33/132 (25%), Positives = 62/132 (46%), Gaps = 1/132 (0%)
Frame = +3
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
++ S++FS DG LA+ D +R +D + + + +LC AWS DGKY+++G
Sbjct: 501 AVLSVAFSPDGRQLASGSGDTTVRFWDLGTQTPMYTCTGHKNWVLCIAWSPDGKYLVSGS 680
Query: 369 EDDLVQVWSMEDRKVVAWG-EGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
+ W + K + GH W++G++ W + N RF S +D
Sbjct: 681 MSGELICWDPQTGKQLGNALTGHKKWITGIS----WEPVHLN----APCRRFVSASKDGD 836
Query: 428 LLLWELEMDEIV 439
+W++ + + +
Sbjct: 837 ARIWDVSLKKCI 872
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 52.8 bits (125), Expect = 4e-07
Identities = 42/135 (31%), Positives = 59/135 (43%), Gaps = 8/135 (5%)
Frame = +1
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFD---YTKEH-----LVCGGKSYYGGLLCCAWS 358
Q I S+ S DG+YL T G D L ++D Y ++ L ++ LL C WS
Sbjct: 820 QDMITSMQLSPDGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWS 999
Query: 359 MDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYR 418
DG + G D +V +W R+++ GHN V+ F PN E I
Sbjct: 1000PDGSKVTAGSSDRMVYIWDTTSRRILYKLPGHNGSVNECVF-----HPN-----EPIV-- 1143
Query: 419 FGSVGQDTQLLLWEL 433
GS D Q+ L E+
Sbjct: 1144-GSCSSDKQIYLGEI 1185
Score = 35.0 bits (79), Expect = 0.076
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 4/85 (4%)
Frame = +1
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
I ++SFS + T G D ++++D K + + + + S DG Y+LT G
Sbjct: 703 ITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQLSPDGSYLLTNGM 882
Query: 370 DDLVQVWSME----DRKVVAWGEGH 390
D + +W M + V EGH
Sbjct: 883 DCKLCIWDMRPYAPQNRCVKILEGH 957
Score = 28.5 bits (62), Expect = 7.2
Identities = 17/70 (24%), Positives = 31/70 (44%)
Frame = +1
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
I TGG D+ V++W + +V +GH ++ + S D +T G
Sbjct: 739 IYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQL--------SPDGSYLLTN-----G 879
Query: 424 QDTQLLLWEL 433
D +L +W++
Sbjct: 880 MDCKLCIWDM 909
Score = 28.1 bits (61), Expect = 9.3
Identities = 28/145 (19%), Positives = 55/145 (37%), Gaps = 8/145 (5%)
Frame = +1
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTK--------EHLVCGGKSYYGGLLCCAWS 358
+ ++ + +++DG + + D LR++D EHL SY CC
Sbjct: 442 KNAVLDLHWTSDGTQIISASPDKTLRLWDTETGKQIKKMVEHL-----SYVNS--CCPTR 600
Query: 359 MDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYR 418
+++G +D ++W M R + D Y + S + Y
Sbjct: 601 RGPPLVVSGSDDGTAKLWDMRQRGSIQ-----------TFPDKYQITAVSFSDASDKIY- 744
Query: 419 FGSVGQDTQLLLWELEMDEIVVPLR 443
+ G D + +W+L E+ + L+
Sbjct: 745 --TGGIDNDVKIWDLRKGEVTMTLQ 813
>TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (50%)
Length = 1227
Score = 52.0 bits (123), Expect = 6e-07
Identities = 26/80 (32%), Positives = 46/80 (57%)
Frame = +2
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
Q S++S+S+S +G L T G + +R +D + + + GL+ CAW GKYIL+
Sbjct: 680 QKSVSSVSWSPNGQELLTCGVEEAVRRWDVSTGKCLQVYEKNGSGLISCAWFPCGKYILS 859
Query: 367 GGEDDLVQVWSMEDRKVVAW 386
G D + +W ++ ++V +W
Sbjct: 860 GLSDKSICMWELDGKEVESW 919
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 50.4 bits (119), Expect = 2e-06
Identities = 25/92 (27%), Positives = 48/92 (52%)
Frame = +1
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S+ + F DG+ A+ G D RV+D V + + +L ++S +G ++ TGG
Sbjct: 1333 SVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLATGG 1512
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
ED+ ++W + +K + H++ +S V F+
Sbjct: 1513 EDNTCRIWDLRKKKSLYTIPAHSNLISQVKFE 1608
Score = 40.8 bits (94), Expect = 0.001
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 1/90 (1%)
Frame = +1
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWS-MDGKYILTG 367
SI ISFS +G +LAT G D R++D K+ + ++ + + +G +++T
Sbjct: 1459 SILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKFEPQEGYFLVTA 1638
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGV 397
D +VWS D K V GH + V+ +
Sbjct: 1639 SYDMTAKVWSSRDFKPVKTLSGHEAKVTSL 1728
Score = 32.3 bits (72), Expect = 0.50
Identities = 41/182 (22%), Positives = 69/182 (37%), Gaps = 3/182 (1%)
Frame = +1
Query: 346 KSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSS 405
K + L A+ GKY+ T D ++W +E + + EGH+ V G+ F S
Sbjct: 1192 KGHLERLARIAFHPSGKYLGTASYDKTWRLWDVETEEELLLQEGHSRSVYGLDFHHDGSL 1371
Query: 406 PNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGS--- 462
S G D +W+L V+ L G + +FS
Sbjct: 1372 A-------------ASCGLDALARVWDLRTGRSVLALE----GHVKSILGISFSPNGYHL 1500
Query: 463 QSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKI 522
+ DN + L+ S+ +P S L++ +V EP ++TA + K+
Sbjct: 1501 ATGGEDNTCRIWDLRKKKSLYTIPAHSNLIS-QVKFEPQEGYF-----LVTASYDMTAKV 1662
Query: 523 WT 524
W+
Sbjct: 1663 WS 1668
Score = 31.2 bits (69), Expect = 1.1
Identities = 15/45 (33%), Positives = 25/45 (55%)
Frame = +1
Query: 355 CAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
C++S DGK + T ++WSM + K V+ +GH + VA+
Sbjct: 967 CSFSRDGKGLATCSFTGATKLWSMPNVKKVSTLKGHTQRATDVAY 1101
>TC84321 GP|22138476|gb|AAM93460.1 hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 748
Score = 45.4 bits (106), Expect = 6e-05
Identities = 26/77 (33%), Positives = 41/77 (52%), Gaps = 8/77 (10%)
Frame = +2
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKE--------HLVCGGKSYYGGLLCCAWSMD 360
SI+S+S S DG L + G D LR + K H + G+ G+ WS D
Sbjct: 482 SISSLSLSPDGRELISAGHDASLRFWSLEKRSCTQEITSHRIMRGE----GVCSVVWSQD 649
Query: 361 GKYILTGGEDDLVQVWS 377
GK++++GG D +V+V++
Sbjct: 650 GKWVVSGGGDGVVKVFA 700
Score = 32.7 bits (73), Expect = 0.38
Identities = 32/153 (20%), Positives = 65/153 (41%), Gaps = 13/153 (8%)
Frame = +2
Query: 242 TCISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--- 297
TCI+ + +FVV+++D + VY+ + + G D TL + +A + +
Sbjct: 116 TCITPLSPNGESFVVSYSDAAILVYDTRTGEQTGTMDSLDTYDGTLLTGVNAVVATTVGL 295
Query: 298 -NPIARWHICQGSIN--------SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSY 348
P + +GS S++ S + + D ++R FD ++
Sbjct: 296 EQPQSGMSEEEGSAGGGPTGGGRSMAGSGVEGVIISGHEDQFVRFFDANSGQCTYNMLAH 475
Query: 349 YGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR 381
+ + S DG+ +++ G D ++ WS+E R
Sbjct: 476 PASISSLSLSPDGRELISAGHDASLRFWSLEKR 574
>TC78591 similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (17%)
Length = 916
Score = 45.1 bits (105), Expect = 7e-05
Identities = 23/80 (28%), Positives = 43/80 (53%)
Frame = +2
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
Q I+S+S+S + L T G + +R +D + + + GL+ C W GK+IL+
Sbjct: 596 QKPISSVSWSPNDEELLTCGVEESIRRWDVSTGKCLQIYEKAGAGLVSCTWFPSGKHILS 775
Query: 367 GGEDDLVQVWSMEDRKVVAW 386
G D + +W ++ ++V +W
Sbjct: 776 GLSDKSICMWELDGKEVESW 835
>TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (59%)
Length = 1824
Score = 42.0 bits (97), Expect = 6e-04
Identities = 25/83 (30%), Positives = 39/83 (46%)
Frame = +2
Query: 350 GGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSN 409
G ++CC +S DGK + + G D V +W+ME K + E H ++ V F PNS
Sbjct: 803 GKVVCCHFSSDGKLLASAGHDKKVVLWNMETLKTQSTPEEHTVIITDVRF-----RPNST 967
Query: 410 DNGETITYRFGSVGQDTQLLLWE 432
+ + DT + LW+
Sbjct: 968 --------QLATSSFDTTVRLWD 1012
>TC78874 similar to PIR|T49098|T49098 hypothetical protein F4F15.300 -
Arabidopsis thaliana, partial (74%)
Length = 1630
Score = 41.2 bits (95), Expect = 0.001
Identities = 17/68 (25%), Positives = 36/68 (52%)
Frame = +2
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
I+FS DG+ A G DG+LR+ ++ ++ + + +S+D +++ + D
Sbjct: 536 ITFSVDGSKFAAGGSDGHLRIMEWPSMRIILDEPRAHKSVRDMDFSLDSEFLASTSTDGS 715
Query: 373 VQVWSMED 380
++W +ED
Sbjct: 716 ARIWKVED 739
>TC87616 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (96%)
Length = 1471
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/95 (27%), Positives = 43/95 (44%), Gaps = 8/95 (8%)
Frame = +3
Query: 292 ARYSKSNPIARWHICQ---GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGK-- 346
A + +NP + + Q SI+S+SFS +L D +R ++ K V
Sbjct: 144 AANTNTNPNKSYEVSQPPTDSISSLSFSPKANFLVATSWDNQVRCWEIAKNGTVVTSTPK 323
Query: 347 ---SYYGGLLCCAWSMDGKYILTGGEDDLVQVWSM 378
S+ +LC AW DG + +GG D ++W +
Sbjct: 324 ASISHDQPVLCSAWKDDGTTVFSGGCDKQAKMWPL 428
>AW691821 similar to GP|10177570|db contains similarity to regulatory protein
Nedd1~gene_id:K18J17.16 {Arabidopsis thaliana}, partial
(19%)
Length = 648
Score = 39.3 bits (90), Expect = 0.004
Identities = 11/39 (28%), Positives = 27/39 (69%)
Frame = +2
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
+Y+ +GG +V++W ++ ++ + W +GH + V+GV ++
Sbjct: 446 RYVCSGGSGQVVKIWDLQKKRCIKWLKGHTNTVTGVMYN 562
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 38.5 bits (88), Expect = 0.007
Identities = 32/143 (22%), Positives = 62/143 (42%), Gaps = 4/143 (2%)
Frame = +1
Query: 272 GAGESSFPILKDQT--LFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDG 329
G GES D+T L+ + + I + H + + +++ A Y T DG
Sbjct: 916 GQGESIITGSADKTVRLWQGSDDGHYNCKQILKDHSAE--VEAVTVHATNNYFVTASLDG 1089
Query: 330 YLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEG 389
++ + + G A+ DG + TG D LV++W ++ + VA +G
Sbjct: 1090 SWCFYELSSGTCLTQVSDSSEGYTSAAFHPDGLILGTGTTDSLVKIWDVKSQANVAKFDG 1269
Query: 390 HNSWVSGVAF--DSYWSSPNSND 410
H V+ ++F + Y+ + ++D
Sbjct: 1270 HVGHVTAISFSENGYYLATAAHD 1338
Score = 31.2 bits (69), Expect = 1.1
Identities = 43/161 (26%), Positives = 64/161 (39%), Gaps = 19/161 (11%)
Frame = +1
Query: 229 HYNKDGILNNSRCTCIS-WVPGGDGAFVVAHADGNLYVYE-----------KNKDGAGES 276
HYN IL + + V + FV A DG+ YE + +G +
Sbjct: 988 HYNCKQILKDHSAEVEAVTVHATNNYFVTASLDGSWCFYELSSGTCLTQVSDSSEGYTSA 1167
Query: 277 SFP----ILKDQTLFSVAHARYSKSNP-IARWHICQGSINSISFSADGAYLATVGRDGYL 331
+F IL T S+ KS +A++ G + +ISFS +G YLAT DG +
Sbjct: 1168 AFHPDGLILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTAISFSENGYYLATAAHDG-V 1344
Query: 332 RVFDYTKEHLVCGGKSYYGGLLCCAWSMD--GKYILTGGED 370
+++D K Y + D G Y+ GG D
Sbjct: 1345 KLWDLRKLKNFRDYGPYDSATPTNSVEFDHSGSYLAVGGSD 1467
>TC87605 homologue to PIR|G96563|G96563 probable coatomer complex subunit
33791-27676 [imported] - Arabidopsis thaliana, partial
(23%)
Length = 765
Score = 38.1 bits (87), Expect = 0.009
Identities = 31/141 (21%), Positives = 56/141 (38%), Gaps = 1/141 (0%)
Frame = +3
Query: 294 YSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLL 353
Y + + + + S F A ++ D ++RV++Y V +++ +
Sbjct: 255 YQSQTMAKSFEVTELPVRSAKFVARKQWVVAGADDMFIRVYNYNTMDKVKVFEAHTDYIR 434
Query: 354 CCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAW-GEGHNSWVSGVAFDSYWSSPNSNDNG 412
C A Y+L+ +D L+++W E + EGH+ +V V F N D
Sbjct: 435 CVAVHPTLPYVLSSSDDMLIKLWDWEKGWICTQIFEGHSHYVMQVTF-------NPKD-- 587
Query: 413 ETITYRFGSVGQDTQLLLWEL 433
T F S D + +W L
Sbjct: 588 ---TNTFASASLDRTIKIWNL 641
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 37.7 bits (86), Expect = 0.012
Identities = 19/81 (23%), Positives = 40/81 (48%), Gaps = 4/81 (4%)
Frame = +2
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYY----GGLLCCAWSMDGKY 363
G +N+++ S DG+ A+ G+DG + ++D + GK Y G ++ +Y
Sbjct: 641 GYVNTVAVSPDGSLCASGGKDGVILLWDLAE------GKRLYSLDAGSIIHALCFSPNRY 802
Query: 364 ILTGGEDDLVQVWSMEDRKVV 384
L + +++W +E + +V
Sbjct: 803 WLCAATESSIKIWDLESKSIV 865
Score = 34.3 bits (77), Expect = 0.13
Identities = 30/126 (23%), Positives = 60/126 (46%), Gaps = 4/126 (3%)
Frame = +2
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKE--HLVCGGKSYYGGLLCCAWSMDGKY--ILTG 367
S++FS D + + RD +++++ E + + G ++ + C +S I++
Sbjct: 389 SVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSA 568
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
D V+VW++ + K+ GH+ +V+ VA SP+ + S G+D
Sbjct: 569 SWDRTVKVWNLTNCKLRNTLAGHSGYVNTVAV-----SPDGS--------LCASGGKDGV 709
Query: 428 LLLWEL 433
+LLW+L
Sbjct: 710 ILLWDL 727
Score = 29.6 bits (65), Expect = 3.2
Identities = 38/166 (22%), Positives = 61/166 (35%), Gaps = 11/166 (6%)
Frame = +2
Query: 322 LATVGRDGYLRVFDYTKEHLVCG-------GKSYYGGLLCCAWSMDGKYILTGGEDDLVQ 374
+ T RD + ++ TKE G G S++ + S DG++ L+G D ++
Sbjct: 152 IVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHF--VQDVVLSSDGQFALSGSWDGELR 325
Query: 375 VWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELE 434
+W + GH V VAF S DN + + S +D + LW
Sbjct: 326 LWDLNAGTSARRFVGHTKDVLSVAF--------SIDNRQIV-----SASRDRTIKLWN-T 463
Query: 435 MDEIVVPLRRGPPGGS----PTFGSPTFSAGSQSSHWDNAVPLGTL 476
+ E ++ G F T S+ WD V + L
Sbjct: 464 LGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNL 601
>TC79131 similar to GP|14031063|gb|AAK52092.1 WD-40 repeat protein
{Lycopersicon esculentum}, partial (71%)
Length = 1224
Score = 37.7 bits (86), Expect = 0.012
Identities = 23/79 (29%), Positives = 38/79 (47%), Gaps = 8/79 (10%)
Frame = +2
Query: 315 FSADGAYLATVGRDGYLRVFDY----TKEHLVCGGKSYY----GGLLCCAWSMDGKYILT 366
FS DG YL + DG++ V+DY K+ L + + +LC +S D + I +
Sbjct: 734 FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIAS 913
Query: 367 GGEDDLVQVWSMEDRKVVA 385
G D ++VW + + A
Sbjct: 914 GSTDGKIKVWRIRTAQCXA 970
>TC87005 similar to GP|9294061|dbj|BAB02018.1 WD40-repeat protein
{Arabidopsis thaliana}, partial (74%)
Length = 1480
Score = 36.6 bits (83), Expect = 0.026
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Frame = +3
Query: 310 INSISFSADGAYLATVGRDGYLRVFD-YTKEHL--VCGGKSYYGGLLCCAWSMDGKYILT 366
+N + +S DG+ T D +FD T E + + + G + +WS DGK +LT
Sbjct: 609 VNCVRYSPDGSKFVTGSSDKKGIIFDGKTGEKIGELSSEGGHTGSIYAVSWSPDGKQVLT 788
Query: 367 GGEDDLVQVWSMEDRKV 383
D +VW + + +
Sbjct: 789 VSADKSAKVWDISEDNI 839
>TC78974 similar to GP|22137238|gb|AAM91464.1 AT5g14050/MUA22_5 {Arabidopsis
thaliana}, partial (40%)
Length = 774
Score = 36.2 bits (82), Expect = 0.034
Identities = 22/79 (27%), Positives = 36/79 (44%)
Frame = +3
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
SI S D +A VG +GY+ + TK + G G + A++ DG+ +L+ G
Sbjct: 33 SIEVFDVSPDSQLIAFVGNEGYILLVS-TKSKQLVGTLKMNGTVRSLAFTEDGQKLLSAG 209
Query: 369 EDDLVQVWSMEDRKVVAWG 387
D + W + R + G
Sbjct: 210 GDGHIYHWDLRTRTCIHKG 266
>TC93203
Length = 681
Score = 35.8 bits (81), Expect = 0.045
Identities = 18/78 (23%), Positives = 37/78 (47%)
Frame = +2
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
+I S+ F+ +G LA DG + +FD + + G ++ + + D I + G
Sbjct: 191 AITSLCFNHNGKILAASAVDGMIHMFDMSAGLQITGWPAHDSSISSILFGPDETSIFSLG 370
Query: 369 EDDLVQVWSMEDRKVVAW 386
D + WS++++ + W
Sbjct: 371 SDGKIFEWSLQNQGQILW 424
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,989,868
Number of Sequences: 36976
Number of extensions: 303513
Number of successful extensions: 1692
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 1511
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1643
length of query: 561
length of database: 9,014,727
effective HSP length: 101
effective length of query: 460
effective length of database: 5,280,151
effective search space: 2428869460
effective search space used: 2428869460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC128638.4


