

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.4 + phase: 0
(301 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
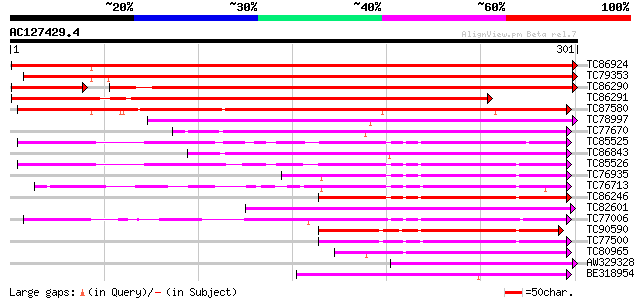
Score E
Sequences producing significant alignments: (bits) Value
TC86924 weakly similar to GP|14422444|dbj|BAB60848. BURP domain-... 595 e-171
TC79353 similar to GP|16588826|gb|AAL26909.1 dehydration-respons... 378 e-105
TC86290 weakly similar to GP|14422444|dbj|BAB60848. BURP domain-... 343 4e-95
TC86291 weakly similar to GP|16588826|gb|AAL26909.1 dehydration-... 342 1e-94
TC87580 similar to GP|18447761|gb|AAL67991.1 dehydration-induced... 310 6e-85
TC78997 weakly similar to PIR|D96529|D96529 probable RAB7 GTP-bi... 167 3e-42
TC77670 similar to PIR|D96529|D96529 probable RAB7 GTP-binding P... 144 4e-35
TC85525 similar to PIR|T08896|T08896 Sali3-2 protein aluminium-... 125 2e-29
TC86843 123 8e-29
TC85526 similar to PIR|T08896|T08896 Sali3-2 protein aluminium-... 119 2e-27
TC76935 similar to SP|P21747|EA92_VICFA Embryonic abundant prote... 114 6e-26
TC76713 similar to PIR|S33622|S33622 ADR6 protein - soybean, par... 112 2e-25
TC86246 weakly similar to PIR|S33622|S33622 ADR6 protein - soybe... 110 9e-25
TC82601 similar to GP|1762584|gb|AAB39546.1| polygalacturonase i... 108 2e-24
TC77006 similar to SP|P21745|EA30_VICFA Embryonic abundant prote... 108 3e-24
TC90590 similar to GP|20914|emb|CAA38756.1|| unknown seed protei... 104 4e-23
TC77500 weakly similar to SP|P21747|EA92_VICFA Embryonic abundan... 102 1e-22
TC80965 similar to PIR|D96529|D96529 probable RAB7 GTP-binding P... 102 1e-22
AW329328 similar to PIR|D96529|D965 probable RAB7 GTP-binding Pr... 93 1e-19
BE318954 similar to PIR|T07587|T07 probable polygalacturonase (E... 90 1e-18
>TC86924 weakly similar to GP|14422444|dbj|BAB60848. BURP domain-containing
protein {Bruguiera gymnorrhiza}, partial (52%)
Length = 1202
Score = 595 bits (1533), Expect = e-171
Identities = 295/306 (96%), Positives = 297/306 (96%), Gaps = 6/306 (1%)
Frame = +1
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP------MDDGKISVDGKIS 55
I + L LALVATHATLPPELYWKSKLPTTQIPKAITDILHP +DDGKISVDGKIS
Sbjct: 85 IFISLSLALVATHATLPPELYWKSKLPTTQIPKAITDILHPRKGVTSVDDGKISVDGKIS 264
Query: 56 VNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPT 115
VNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPT
Sbjct: 265 VNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPT 444
Query: 116 FLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLE 175
FLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLE
Sbjct: 445 FLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLE 624
Query: 176 SMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCH 235
SMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCH
Sbjct: 625 SMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCH 804
Query: 236 KTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQ 295
KTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQ
Sbjct: 805 KTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQ 984
Query: 296 VVWFSK 301
VVWFSK
Sbjct: 985 VVWFSK 1002
>TC79353 similar to GP|16588826|gb|AAL26909.1 dehydration-responsive protein
RD22 {Prunus persica}, partial (64%)
Length = 1324
Score = 378 bits (971), Expect = e-105
Identities = 195/312 (62%), Positives = 229/312 (72%), Gaps = 18/312 (5%)
Frame = +2
Query: 8 LALVATHAT-LPPELYWKSKLPTTQIPKAITDILHP-----------MDDGKISVD---- 51
L + AT+A LPP+LYWKS LP + +PKAIT++LHP +D GK VD
Sbjct: 104 LVVGATNAVMLPPQLYWKSMLPNSPMPKAITNLLHPAGYWSEEKGTWVDVGKGGVDVGVR 283
Query: 52 -GKISVNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTS 110
G G V + Y A+E +LHD N+A+F LEKDLHHGTK N+QF+KT+
Sbjct: 284 KGYYEGGGTDVNVGVGRSPFIYNYAASETQLHDKPNVALFFLEKDLHHGTKLNLQFSKTT 463
Query: 111 DHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLC 170
+ TFLP VANSIPFSSNK+E I+N +IK+GS +IVKNTISECE GIKGEEK+C
Sbjct: 464 SNAATFLPRQVANSIPFSSNKMEYIINKLNIKKGSKGVQIVKNTISECEEQGIKGEEKVC 643
Query: 171 VTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGE-NNLVVCHKRNYPY 229
VTSLESM+DFTT KLG NV+ VSTEVN ES LQQY IA+GVKK+GE N VVCHK NYPY
Sbjct: 644 VTSLESMVDFTTSKLGKNVEAVSTEVNKESNLQQYTIASGVKKLGEKNKAVVCHKENYPY 823
Query: 230 AVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCH 289
AVFYCHKTD TK YSVPLEGADGSRVKA+A+CH+DTS+W+ KHLAFQVLKVQ GT PVCH
Sbjct: 824 AVFYCHKTDTTKAYSVPLEGADGSRVKAIAVCHTDTSEWNPKHLAFQVLKVQPGTVPVCH 1003
Query: 290 ILQQGQVVWFSK 301
+L + VVW K
Sbjct: 1004LLPEDHVVWIRK 1039
>TC86290 weakly similar to GP|14422444|dbj|BAB60848. BURP domain-containing
protein {Bruguiera gymnorrhiza}, partial (87%)
Length = 1302
Score = 343 bits (881), Expect = 4e-95
Identities = 176/251 (70%), Positives = 201/251 (79%), Gaps = 3/251 (1%)
Frame = +2
Query: 54 ISVNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKT-SDH 112
+SV K+ +Y+ A A+E +LHD N+A+F LEKDLH GTK N+QFTKT S+
Sbjct: 500 VSVGNKSPFLYNYA--------ASETQLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSND 655
Query: 113 GPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVT 172
FLP +VANSIPFSS+K+ENILN FSIK+G+ ESEIVKNTISECE GIKGEEKLCVT
Sbjct: 656 DAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVT 835
Query: 173 SLESMIDFTTLKLGNNVDTVSTEVNGESG-LQQYVIANGVKKMGENN-LVVCHKRNYPYA 230
SLESM+DFTT KLGNNV+ VSTEV ES LQ+YV+A GVKK+GE N VVCHK +YPYA
Sbjct: 836 SLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYA 1015
Query: 231 VFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHI 290
VFYCHKTD+TKVYSVPLEG DGSRVKAVA+CH+DTSQW+ KHLAFQVL VQ GT PVCH
Sbjct: 1016VFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHF 1195
Query: 291 LQQGQVVWFSK 301
L Q VVW SK
Sbjct: 1196LPQDHVVWVSK 1228
Score = 72.8 bits (177), Expect = 2e-13
Identities = 33/40 (82%), Positives = 37/40 (92%)
Frame = +2
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILH 41
I +VL LALVATHA LPPE+YWKSKLPTTQ+PKAITD+LH
Sbjct: 95 IFIVLNLALVATHAALPPEVYWKSKLPTTQMPKAITDLLH 214
>TC86291 weakly similar to GP|16588826|gb|AAL26909.1 dehydration-responsive
protein RD22 {Prunus persica}, partial (42%)
Length = 829
Score = 342 bits (877), Expect = 1e-94
Identities = 171/255 (67%), Positives = 197/255 (77%)
Frame = +1
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTR 61
I + L LALVATHA LPPELYWKSKLPTTQIPKAITDILHP G + V R
Sbjct: 85 IFISLNLALVATHAALPPELYWKSKLPTTQIPKAITDILHPRKGGTW-----VDVGNAGR 249
Query: 62 IVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDV 121
+ ++ DA++ +LHD RNLA+F EKDLH+GTK N+QFTKTSD+G TFLP +V
Sbjct: 250 V--QQLLFIYWPYDASDTQLHDYRNLALFFFEKDLHNGTKLNMQFTKTSDYGATFLPREV 423
Query: 122 ANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFT 181
ANSIPFSSNK+ENILNYFSIKQGS E EIVKNTI CE I+GEEK CVTSLESM+DFT
Sbjct: 424 ANSIPFSSNKVENILNYFSIKQGSAEYEIVKNTIGSCEMPAIEGEEKSCVTSLESMVDFT 603
Query: 182 TLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
T KLGNNV+ VSTEVN ES +QQY+IA GVKK+GEN +VVCH +YPY VFYCHK ATK
Sbjct: 604 TSKLGNNVEAVSTEVNKESDIQQYIIAKGVKKLGENKIVVCHPMDYPYTVFYCHKLRATK 783
Query: 242 VYSVPLEGADGSRVK 256
VY +P+EG DG ++K
Sbjct: 784 VYYLPMEGVDGPKIK 828
>TC87580 similar to GP|18447761|gb|AAL67991.1 dehydration-induced protein
RD22-like protein {Gossypium hirsutum}, partial (76%)
Length = 1290
Score = 310 bits (793), Expect = 6e-85
Identities = 168/311 (54%), Positives = 217/311 (69%), Gaps = 17/311 (5%)
Frame = +2
Query: 5 VLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISVD-GK-ISVN--- 57
+L L LVATHA LPPELYWKS LPTT +PKAITDILHP M+D +V GK +SVN
Sbjct: 107 ILNLVLVATHAALPPELYWKSVLPTTPMPKAITDILHPGWMEDKSTNVGVGKGVSVNAGH 286
Query: 58 ---GK--TRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDH 112
GK T +V +P + Y A E +LHD+ A+F LEKDLH GTK ++ F ++S+
Sbjct: 287 KRKGKPATVVVGPHSP-FAYNYAATETQLHDDPRAALFFLEKDLHPGTKMDLNFIRSSNV 463
Query: 113 GPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVT 172
TFLP VA++ PFSS KL +I N FS+K S E+ ++KNTI ECE I+GEEK C T
Sbjct: 464 A-TFLPRQVASATPFSSEKLSHIFNKFSVKPESEEAHVMKNTIEECEDAAIQGEEKYCAT 640
Query: 173 SLESMIDFTTLKLGNNVDTVSTEV--NGESGLQQYVIANGVKKM-GENNLVVCHKRNYPY 229
SLESM+DF+ KLG V VST V N +GL++Y + +G+ K+ E+ VVCHK+NYPY
Sbjct: 641 SLESMVDFSISKLGKRVKVVSTTVVDNKSAGLKKYTLKSGLMKLTAEDKAVVCHKQNYPY 820
Query: 230 AVFYCHKTDATKVYSVPLEGADGSRVK--AVAICHSDTSQWSLKHLAFQVLKVQQGTFPV 287
A+FYCHKT++T+ Y VPLE +G+RVK AV ICH+DTS+W+ +H+AFQVLK++ GT V
Sbjct: 821 AIFYCHKTESTRAYFVPLEADNGTRVKADAVVICHTDTSKWNPEHIAFQVLKIKPGTLSV 1000
Query: 288 CHILQQGQVVW 298
CH L +VW
Sbjct: 1001CHFLPVDHIVW 1033
>TC78997 weakly similar to PIR|D96529|D96529 probable RAB7 GTP-binding
Protein [imported] - Arabidopsis thaliana, partial (27%)
Length = 1039
Score = 167 bits (424), Expect = 3e-42
Identities = 92/234 (39%), Positives = 128/234 (54%), Gaps = 6/234 (2%)
Frame = +2
Query: 74 NDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDH-GPTFLPGDVANSIPFSSNKL 132
+D N + + +L +F KDL G I F K P A S+PFSSN+L
Sbjct: 128 HDHNHMSHNIDPSLMVFFTLKDLKVGNIMQIYFPKRDPSTSPKLWSKQKAESLPFSSNQL 307
Query: 133 ENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVD-- 190
+L +FS Q S ++ +KNT+ ECE+ IKGE KLC TS ESM++FT LG+ +
Sbjct: 308 SYLLKFFSFSQDSPQAMAMKNTLRECESKPIKGEVKLCATSFESMLEFTQNVLGSKYEIQ 487
Query: 191 --TVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDA-TKVYSVPL 247
+ LQ Y I +K++ +V CH YPYAVFYCH ++ +VY L
Sbjct: 488 GYATLHKTKSSVTLQNYTIVEILKEILAPKMVACHTVPYPYAVFYCHGQESDNRVYRASL 667
Query: 248 EGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
G +G +V+A+A+CH DTSQW+ H++FQVL+V GT VCH +W K
Sbjct: 668 VGENGDKVEAMAVCHMDTSQWAPSHVSFQVLEVTPGTSSVCHFFPADNYIWVPK 829
>TC77670 similar to PIR|D96529|D96529 probable RAB7 GTP-binding Protein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 1649
Score = 144 bits (363), Expect = 4e-35
Identities = 84/217 (38%), Positives = 117/217 (53%), Gaps = 5/217 (2%)
Frame = +2
Query: 87 LAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGST 146
LA F ++ DL+ G +QF FLP VA+ IPFS ++L ++L FS+ + S
Sbjct: 671 LAFFTMD-DLYVGNVMTLQFPVREY--ANFLPKKVADYIPFSKSQLPSLLQLFSLTKDSP 841
Query: 147 ESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN----NVDTVSTEVNGESGL 202
+ E +K+ I +CE KGE K C TSLESM++F LG N+ T S + L
Sbjct: 842 QGEDMKDIIDQCEFEPTKGETKACPTSLESMLEFVHSVLGTEARYNIHTTSYPTTSGARL 1021
Query: 203 QQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTD-ATKVYSVPLEGADGSRVKAVAIC 261
Q Y I K + V CH R YPYA++YCH D +KV+ V L+G G + A+ IC
Sbjct: 1022QNYTILKISKDIYAPKWVACHPRPYPYALYYCHYLDIGSKVFKVLLKGQYGDIMDALGIC 1201
Query: 262 HSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
H DTS + H FQ+L ++ G P+CH V+W
Sbjct: 1202HLDTSDMNPNHFIFQLLGIKPGEAPMCHFFPVKHVLW 1312
>TC85525 similar to PIR|T08896|T08896 Sali3-2 protein aluminium-induced -
soybean, partial (89%)
Length = 1076
Score = 125 bits (314), Expect = 2e-29
Identities = 92/296 (31%), Positives = 145/296 (48%), Gaps = 2/296 (0%)
Frame = +3
Query: 5 VLQLALVA-THATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIV 63
VL LAL +HA+LP E YW++ P T IP ++ ++L P +G
Sbjct: 78 VLSLALAGGSHASLPEEEYWEAVWPNTPIPSSLRELLKPGPEG----------------- 206
Query: 64 YDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVAN 123
E D E+ D + F E +L+ G +QF+K P P V
Sbjct: 207 --------VEIDDLPMEVDDTQYPKTFFYEHELYPGKTMKVQFSKR----PFAQPYGVYT 350
Query: 124 SIPFSSNKLENILNYFSIKQGSTESEI-VKNTISECEAYGIKGEEKLCVTSLESMIDFTT 182
+ ++++I K+G T +E+ VK +E GE+K C SL ++I F+
Sbjct: 351 WM----REIKDIE-----KEGYTFNEVCVKKAAAE-------GEQKFCAKSLGTLIGFSI 482
Query: 183 LKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKV 242
KLG N+ +S+ + +QY I + V+ +GE V+CH+ N+ VFYCH+ T
Sbjct: 483 SKLGKNIQALSSSFIDKH--EQYKIES-VQNLGEK-AVMCHRLNFQKVVFYCHEIHGTTA 650
Query: 243 YSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ VPL DG + +A+A+CH+DTS + + L Q++K G+ PVCH L ++W
Sbjct: 651 FMVPLVANDGRKTQALAVCHTDTSGMNHEMLQ-QIMKADPGSKPVCHFLGNKAILW 815
>TC86843
Length = 1157
Score = 123 bits (309), Expect = 8e-29
Identities = 72/211 (34%), Positives = 111/211 (52%), Gaps = 7/211 (3%)
Frame = +1
Query: 95 DLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNT 154
DL+ G +QF FL A+SIP + ++L ++L SI + S +++ + N+
Sbjct: 319 DLYVGNVMTLQFPVQVVS--PFLSKKEADSIPLAMSQLPSVLQLLSIPEDSPQAKSMINS 492
Query: 155 ISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGES----GLQQYVIANG 210
+ ECE+ I GE K C SLESM++F G++ N S LQ+Y I
Sbjct: 493 LGECESETITGETKTCANSLESMLEFVNTIFGSDAKISILTTNKPSPTAVPLQKYTILEV 672
Query: 211 VKKMGENNLVVCHKRNYPYAVFYCHK-TDATKVYSVPLEG-ADGSRVKAVAICHSDTSQW 268
+ V CH +YPYA+++CH T+V+ V L G +G +++AV ICH DTS W
Sbjct: 673 SHDIDAPKWVACHPLSYPYAIYFCHYIATGTRVFKVSLVGDENGDKMEAVGICHLDTSDW 852
Query: 269 SLKHLAFQVLKVQQG-TFPVCHILQQGQVVW 298
+ H+ F+ L ++ G PVCH L ++W
Sbjct: 853 NPDHIIFRQLSIKAGKNSPVCHFLPVNHLLW 945
>TC85526 similar to PIR|T08896|T08896 Sali3-2 protein aluminium-induced -
soybean, partial (89%)
Length = 1095
Score = 119 bits (297), Expect = 2e-27
Identities = 88/296 (29%), Positives = 140/296 (46%), Gaps = 2/296 (0%)
Frame = +2
Query: 5 VLQLALVA-THATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIV 63
VL LAL +HA++P E YW++ P T IP ++ ++L P G
Sbjct: 74 VLSLALAGGSHASVPEEEYWEAVWPNTPIPTSLLELLKPGPKG----------------- 202
Query: 64 YDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVAN 123
E D E+ D + F E +L+ G N+QF+K P
Sbjct: 203 --------VEIDDLPTEIDDTQFPTNFFYEHELYPGKTMNMQFSKRPLAQPY-------- 334
Query: 124 SIPFSSNKLENILNYFSIKQGSTESEI-VKNTISECEAYGIKGEEKLCVTSLESMIDFTT 182
+ F + ++++ K+G T E+ VKN K EEK C SL ++I F
Sbjct: 335 GVYFWMHDIKDLQ-----KEGYTIDEMCVKNKPK-------KVEEKFCAKSLGTLIGFAI 478
Query: 183 LKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKV 242
KLG N+ ++S+ + +QY I + V+ +G+ V+CH+ N+ VFYCH+ T
Sbjct: 479 SKLGKNIQSLSSSFIDKH--EQYKIES-VQNLGDK-AVMCHRLNFQKVVFYCHEVHGTTA 646
Query: 243 YSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ VPL DG++ A+A CH+D S + +H+ Q++K G+ VCH L ++W
Sbjct: 647 FKVPLVANDGTKTHAIATCHADISGMN-QHMLHQIMKGDPGSNHVCHFLGNKAILW 811
>TC76935 similar to SP|P21747|EA92_VICFA Embryonic abundant protein USP92
precursor. [Broad bean] {Vicia faba}, partial (40%)
Length = 1840
Score = 114 bits (284), Expect = 6e-26
Identities = 64/163 (39%), Positives = 92/163 (56%), Gaps = 9/163 (5%)
Frame = +1
Query: 145 STESEIVKNTISECEAYGIK---------GEEKLCVTSLESMIDFTTLKLGNNVDTVSTE 195
S V +T E EA+ ++ GE+K C SLESM+DF KLG N+ +S+
Sbjct: 1075 SQHQHFVTHTSDENEAHILEDYCGRPSAIGEDKHCAPSLESMMDFAISKLGKNIKVMSSS 1254
Query: 196 VNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRV 255
+ QYV+ V K+G+N V+CH+ N+ +FYCH+ +AT Y VPL +DG++
Sbjct: 1255 FSKNH--DQYVVEE-VNKIGDN-AVMCHRLNFKKVLFYCHQVNATTTYMVPLVASDGTKS 1422
Query: 256 KAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
KA+ ICH DT ++ + VLKV+ GT PVCH + V W
Sbjct: 1423 KALTICHHDTRGMD-PNVLYDVLKVKPGTVPVCHFIGNKAVAW 1548
Score = 36.6 bits (83), Expect = 0.012
Identities = 17/46 (36%), Positives = 26/46 (55%)
Frame = +1
Query: 8 LALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGK 53
LALV + + E YWKS P T +PK+++D+L + + GK
Sbjct: 67 LALVGINGSKSGEEYWKSVWPNTLMPKSLSDLLLSESGTSVPIQGK 204
>TC76713 similar to PIR|S33622|S33622 ADR6 protein - soybean, partial (67%)
Length = 1790
Score = 112 bits (280), Expect = 2e-25
Identities = 84/297 (28%), Positives = 136/297 (45%), Gaps = 12/297 (4%)
Frame = +2
Query: 14 HATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAAPIYFYE 73
H +LP + YW++ P T IP A+ D+L P +G D P+ F +
Sbjct: 755 HGSLPQK-YWEAVWPNTPIPIAMQDLLKPEHEGDEIED---------------LPMEFEK 886
Query: 74 NDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLE 133
E N +L+ G N+QF K+ PF+
Sbjct: 887 QQYPEDLFFQN----------ELYPGNTMNLQFDKS----------------PFAQPA-- 982
Query: 134 NILNYFSIKQGSTESEIVKNTISECEAYGIK----------GEEKLCVTSLESMIDFTTL 183
++ Y ++ +KNT E EAY + GE+K C SL S+I+F
Sbjct: 983 GLIKYLGA------NDDIKNT--EKEAYNVDELCVKKRATIGEQKHCAKSLGSLIEFAIS 1138
Query: 184 KLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVY 243
KLGNN+ +S+ + +QY + + V+ +G+ V+CH+ N+ V+YCH+ AT +
Sbjct: 1139KLGNNIQALSSSLIDNQ--EQYTVES-VQNLGDKG-VMCHRLNFQKVVYYCHRIRATTAF 1306
Query: 244 SVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQG--TFPVCHILQQGQVVW 298
VPL DG++ +A+A+CH+DTS + + + LK+ +PVCH L ++W
Sbjct: 1307MVPLVAGDGTKTQAIAVCHADTSGMN-QQMLHDALKLDSADIKYPVCHFLGNKAIMW 1474
>TC86246 weakly similar to PIR|S33622|S33622 ADR6 protein - soybean, partial
(56%)
Length = 1324
Score = 110 bits (274), Expect = 9e-25
Identities = 56/134 (41%), Positives = 82/134 (60%)
Frame = +2
Query: 165 GEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHK 224
GEEK C SLESM+DF KLG N+ +S+ + QY++ V+K+G+N V+CH+
Sbjct: 689 GEEKHCAYSLESMMDFAISKLGKNIKVMSSSFSQSQ--DQYMVEE-VRKIGDN-AVMCHR 856
Query: 225 RNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGT 284
N FYCH+ +AT Y VPL +DG++ KA+ ICH DT ++ ++VL+V+ GT
Sbjct: 857 MNLKKVGFYCHQINATTTYMVPLVASDGTKSKALTICHHDTRGMD-PNMLYEVLQVKPGT 1033
Query: 285 FPVCHILQQGQVVW 298
PVCH + + W
Sbjct: 1034VPVCHFIGNKAIAW 1075
>TC82601 similar to GP|1762584|gb|AAB39546.1| polygalacturonase isoenzyme 1
beta subunit homolog {Arabidopsis thaliana}, partial
(32%)
Length = 1072
Score = 108 bits (271), Expect = 2e-24
Identities = 63/178 (35%), Positives = 89/178 (49%), Gaps = 3/178 (1%)
Frame = +2
Query: 126 PFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKL 185
P S+KL + F + + S+ ++++ + +SECE GE K CV SLE MIDF T L
Sbjct: 281 PLHSSKLNEMRQVFKVSENSSMNKMMVDALSECERAPSVGETKRCVGSLEDMIDFATSVL 460
Query: 186 GNNVDTVSTE-VNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYS 244
G++V STE VNG G+ V CH+ +PY ++YCH VY
Sbjct: 461 GHSVTVRSTENVNGAGKDVMVGKVKGINGGKVTESVSCHQSLFPYLLYYCHSVPNVHVYE 640
Query: 245 VP-LEGADGSRVK-AVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFS 300
L+ +++ VAICH DT+ WS H AF L G VCH + + + W S
Sbjct: 641 ADLLDPVSKAKINHGVAICHLDTTAWSSTHGAFMALGSAPGQIEVCHWIFENDMTWSS 814
>TC77006 similar to SP|P21745|EA30_VICFA Embryonic abundant protein VF30.1
precursor. [Broad bean] {Vicia faba}, complete
Length = 1136
Score = 108 bits (270), Expect = 3e-24
Identities = 89/298 (29%), Positives = 130/298 (42%), Gaps = 7/298 (2%)
Frame = +2
Query: 8 LALVATHAT-LPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDA 66
LA V AT L E YW++ P T +PKA +D+L P GKT +
Sbjct: 68 LAFVGISATTLSSEDYWQTIWPNTPLPKAFSDLLLPY--------------GKTNNL--- 196
Query: 67 APIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIP 126
PI + + + E DL+ G K I
Sbjct: 197 -PI----------KTEELNQYSTLFFEHDLYPGKKM----------------------IL 277
Query: 127 FSSNKLENILNYFSI-KQGSTESEIVKNTISE-----CEAYGIKGEEKLCVTSLESMIDF 180
++N L N + F +QG T+S + N + C + GE K CV+SLESM++
Sbjct: 278 GNNNSLRNTVRPFGKPRQGITDSIWLANKERQSLDDFCNSPTATGERKHCVSSLESMVEH 457
Query: 181 TTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDAT 240
G + + G + QYV+ VKK+G+N V+CH+ N+ VF CH+ AT
Sbjct: 458 VISHFGTSKIKAISSTFGVNQ-DQYVVEE-VKKVGDN-AVMCHRLNFEKVVFNCHQVQAT 628
Query: 241 KVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
Y V L DG + KA+ +CH DT + + L ++ LKV+ GT P+CH + W
Sbjct: 629 TAYVVSLVAPDGGKAKALTVCHHDTRGMNAE-LLYEALKVEPGTIPICHFIGNKAAAW 799
>TC90590 similar to GP|20914|emb|CAA38756.1|| unknown seed protein {Pisum
sativum}, partial (44%)
Length = 675
Score = 104 bits (260), Expect = 4e-23
Identities = 56/131 (42%), Positives = 83/131 (62%), Gaps = 1/131 (0%)
Frame = +1
Query: 165 GEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHK 224
GEEK C SL+SM++F KLG N+ +S+ QY + + VKK+G+ V+CH+
Sbjct: 238 GEEKYCALSLKSMMNFAISKLGTNIKVISSSF--AQNQDQYKV-DEVKKIGDK-AVMCHR 405
Query: 225 RNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGT 284
N+ VFYCH+ +AT VY V L +DG++ KA+ ICH DT + + +++LKV+ GT
Sbjct: 406 LNFKDMVFYCHQVNATTVYMVLLVASDGTKAKALTICHHDTRGMN-PDVLYELLKVKPGT 582
Query: 285 FPVCH-ILQQG 294
P+CH + QQG
Sbjct: 583 IPICHFVWQQG 615
>TC77500 weakly similar to SP|P21747|EA92_VICFA Embryonic abundant protein
USP92 precursor. [Broad bean] {Vicia faba}, partial
(40%)
Length = 1128
Score = 102 bits (255), Expect = 1e-22
Identities = 56/134 (41%), Positives = 78/134 (57%)
Frame = +3
Query: 165 GEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHK 224
GE+K C TS ESM +F KLG + + S QYV+ V+K+ + V+CH+
Sbjct: 306 GEDKFCATSSESMKEFAISKLGAKIKSYSGYF--AKNQDQYVVEE-VRKIADKG-VMCHR 473
Query: 225 RNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGT 284
N+ VFYCH+ +A+ Y VPL +DGS+V A+A CH DT + +L +VLKV+ GT
Sbjct: 474 MNFEKVVFYCHQVNASTTYMVPLLASDGSKVNALAACHHDTRGMN-PNLLDEVLKVKPGT 650
Query: 285 FPVCHILQQGQVVW 298
PVCH + V W
Sbjct: 651 IPVCHFVGNKAVAW 692
>TC80965 similar to PIR|D96529|D96529 probable RAB7 GTP-binding Protein
[imported] - Arabidopsis thaliana, partial (14%)
Length = 533
Score = 102 bits (255), Expect = 1e-22
Identities = 55/131 (41%), Positives = 71/131 (53%), Gaps = 5/131 (3%)
Frame = +1
Query: 173 SLESMIDFTTLKLGNN----VDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYP 228
SLES+ DF G N V T + LQ Y I+ V ++ N V CH YP
Sbjct: 7 SLESLFDFANYMFGPNSKFKVLTTTHVTKSSIPLQNYTISK-VNEISVPNAVGCHPMPYP 183
Query: 229 YAVFYCHKTDA-TKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPV 287
+AVFYCH T +Y + +EG +G V+A AICH DTS+W H+AF+VL V+ G PV
Sbjct: 184 FAVFYCHSQKGDTSLYEIVVEGENGGIVQAAAICHMDTSKWDADHVAFRVLNVKPGNSPV 363
Query: 288 CHILQQGQVVW 298
CH +VW
Sbjct: 364 CHFSPPDNLVW 396
>AW329328 similar to PIR|D96529|D965 probable RAB7 GTP-binding Protein
[imported] - Arabidopsis thaliana, partial (6%)
Length = 591
Score = 92.8 bits (229), Expect = 1e-19
Identities = 42/100 (42%), Positives = 59/100 (59%), Gaps = 1/100 (1%)
Frame = +1
Query: 203 QQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDA-TKVYSVPLEGADGSRVKAVAIC 261
Q Y I + ++ +V CH YPYAVFYCH ++ +VY V L G +G +V+A+ +C
Sbjct: 67 QNYTIVEIMMEIRAPKMVACHTVPYPYAVFYCHSQESENRVYKVLLGGENGDKVEAMVVC 246
Query: 262 HSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
H DTSQW+ H++FQVL V G+ VCH +W K
Sbjct: 247 HLDTSQWAPSHVSFQVLGVTPGSSSVCHFFPADNYIWVPK 366
>BE318954 similar to PIR|T07587|T07 probable polygalacturonase (EC 3.2.1.15)
1 - tomato, partial (23%)
Length = 556
Score = 89.7 bits (221), Expect = 1e-18
Identities = 53/150 (35%), Positives = 72/150 (47%), Gaps = 4/150 (2%)
Frame = +2
Query: 153 NTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTE-VNGESGLQQYVIANGV 211
+++ ECE +GE K CV S+E MIDF LG NV +TE VNG +G+
Sbjct: 5 DSLGECERVPARGETKKCVGSIEDMIDFAASVLGRNVVVRTTENVNGSKKDLMVGHVSGI 184
Query: 212 KKMGE-NNLVVCHKRNYPYAVFYCHKTDATKVYSVPL--EGADGSRVKAVAICHSDTSQW 268
G+ V CH+ + Y ++YCH +VY L + VA+CH DTS W
Sbjct: 185 NNGGKMTRTVSCHQSLFQYLLYYCHSVPKVRVYQAELLDPKIKDKINQGVAVCHLDTSDW 364
Query: 269 SLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
S H AF L G VCH + + + W
Sbjct: 365 SPTHGAFVSLGSGPGQIEVCHWIFENDMSW 454
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,286,452
Number of Sequences: 36976
Number of extensions: 117112
Number of successful extensions: 690
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 630
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 638
length of query: 301
length of database: 9,014,727
effective HSP length: 96
effective length of query: 205
effective length of database: 5,465,031
effective search space: 1120331355
effective search space used: 1120331355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC127429.4


