

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127428.4 + phase: 0
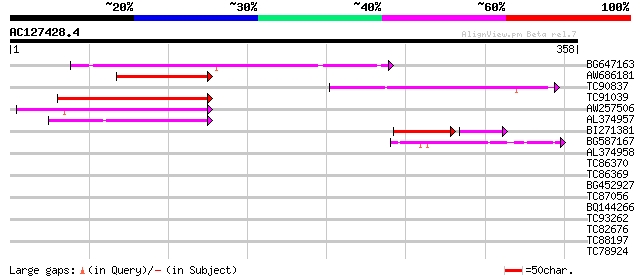
(358 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.... 132 3e-31
AW686181 129 2e-30
TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical... 105 4e-23
TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24... 96 2e-20
AW257506 weakly similar to GP|9757943|dbj| emb|CAB72466.1~gene_i... 60 1e-09
AL374957 55 3e-08
BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa... 54 3e-08
BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.... 44 1e-04
AL374958 GP|8979817|emb| dJ447E21.2 (Similar to X.laevis Xrel2 p... 34 0.076
TC86370 similar to GP|15451010|gb|AAK96776.1 Unknown protein {Ar... 33 0.22
TC86369 similar to GP|15451010|gb|AAK96776.1 Unknown protein {Ar... 31 0.64
BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH o... 29 3.2
TC87056 weakly similar to GP|20521237|dbj|BAB91753. contains EST... 28 7.1
BQ144266 similar to GP|19887436|gb| Fe-S oxidoreductase family p... 28 7.1
TC93262 weakly similar to PIR|T07972|T07972 leucoanthocyanidin d... 27 9.3
TC82676 similar to GP|8843849|dbj|BAA97375.1 contains similarity... 27 9.3
TC88197 homologue to GP|6739629|gb|AAF27340.1| abscisic acid-act... 27 9.3
TC78924 similar to GP|20197503|gb|AAC95160.2 putative translatio... 27 9.3
>BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.8~similar to
unknown protein {Arabidopsis thaliana}, partial (51%)
Length = 744
Score = 132 bits (331), Expect = 3e-31
Identities = 83/249 (33%), Positives = 123/249 (49%), Gaps = 45/249 (18%)
Frame = +3
Query: 39 LCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDL-KSSKHMGV 97
L K P R+ LT + +E+L G RC E FRM+K +F+KLC L L + + + +
Sbjct: 3 LYKTPPRSYNLT--KFFEEVLNGPNQRCLENFRMDKVVFYKLCDILETKGLLRDTNRIKI 176
Query: 98 EEMVAMFLVVVGHGVGNRMIQERFQHSGETVR---------------------------- 129
EE +A+FL ++GH + R +QE F +SGET+
Sbjct: 177 EEQLAIFLFIIGHNLRIRGVQELFHYSGETISRHFNNVLNAVMSISKEYFQPPGEDVASM 356
Query: 130 ----------------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVL 173
IDG +V V EQ F + G +QN++A C +++ F +VL
Sbjct: 357 IAEDDRFFPYFKDCVGAIDGIYVPVTVGVDEQGPFRNKDGLLSQNVLAACSFDLKFCYVL 536
Query: 174 AGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFR 233
AGWEG+A + +VF+ A+T N P+GKYYLVD+ +P G+I PY YH EF
Sbjct: 537 AGWEGSASNLQVFNSAITRKNK--LQVPEGKYYLVDNKFPNVPGFIAPYPRTPYHSKEF- 707
Query: 234 RSSGFENHN 242
+G++ N
Sbjct: 708 -PTGYQPQN 731
>AW686181
Length = 304
Score = 129 bits (323), Expect = 2e-30
Identities = 61/61 (100%), Positives = 61/61 (100%)
Frame = +3
Query: 68 EMFRMEKHIFHKLCHELVEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGET 127
EMFRMEKHIFHKLCHELVEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGET
Sbjct: 3 EMFRMEKHIFHKLCHELVEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGET 182
Query: 128 V 128
V
Sbjct: 183 V 185
>TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical
protein~similar to Oryza sativa chromosome10
OSJNBa0079H13.17, partial (7%)
Length = 596
Score = 105 bits (261), Expect = 4e-23
Identities = 58/151 (38%), Positives = 84/151 (55%), Gaps = 6/151 (3%)
Frame = +1
Query: 203 GKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGV 262
G+YYLVD GYP GY+ PY RYH +F N E FN HSSLR IER+FGV
Sbjct: 4 GRYYLVDKGYPDKEGYMVPYPRIRYHQSQFEHEPP-TNAQEAFNRAHSSLRSCIERSFGV 180
Query: 263 WKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHD---- 318
K R+ IL MP+F +TQ+ +++A A+HN+IR +++ D F + E I +D
Sbjct: 181 LKKRWKILNKMPQFSVKTQIDVIIAAFALHNYIRINSQDDAMFTILERHPNYIPNDELPD 360
Query: 319 --DDHRSTNLNQSQSFNVASSSEMDHARNSI 347
D ++ + + +S + EM RN++
Sbjct: 361 IVDGYQGSERQEGRSGRSTKTKEM---RNNV 444
>TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24.14
[imported] - Arabidopsis thaliana, partial (4%)
Length = 732
Score = 96.3 bits (238), Expect = 2e-20
Identities = 46/99 (46%), Positives = 63/99 (63%), Gaps = 1/99 (1%)
Frame = +1
Query: 31 LGEYATKYLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHEL-VEHDL 89
+ Y KY+ KEPC TS G W+ EIL G+P RC FRM+ +F +LC +L ++ L
Sbjct: 172 IATYYYKYIYKEPCMTSLQRGQDWMNEILNGHPVRCMNAFRMDPTLFKQLCEDLQSKYGL 351
Query: 90 KSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETV 128
+ SK M VEE V +F+ + G NR ++ERFQHSGET+
Sbjct: 352 QPSKRMTVEEKVGIFVYTLAMGASNRDVRERFQHSGETI 468
>AW257506 weakly similar to GP|9757943|dbj|
emb|CAB72466.1~gene_id:MJC20.8~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 510
Score = 60.5 bits (145), Expect = 1e-09
Identities = 38/128 (29%), Positives = 59/128 (45%), Gaps = 4/128 (3%)
Frame = +2
Query: 5 KEQLEHNTQEEEDDDTFEESYILAALLGE---YATKYLCKEPCRTSELTGHAWVQEILQG 61
K + + E ++ TF + LLG Y Y KEP R EL ++ + +G
Sbjct: 14 KRKRDEEEMRELEETTFIVVSVATMLLGAMIWYYNTYFVKEPARNLELERRNFLNRLYRG 193
Query: 62 NPTRCYEMFRMEKHIFHKLCHELVEH-DLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQER 120
C E R+ K F KLC L E L ++++ E VAMFL ++ H + R++
Sbjct: 194 TEADCIEQLRVSKRTFFKLCKILKEKGQLVRTRNVPTTEAVAMFLHILAHNLKYRVVHFS 373
Query: 121 FQHSGETV 128
+ S ET+
Sbjct: 374 YCRSKETI 397
>AL374957
Length = 490
Score = 55.5 bits (132), Expect = 3e-08
Identities = 41/105 (39%), Positives = 50/105 (47%), Gaps = 1/105 (0%)
Frame = +1
Query: 25 YILAALLGEYATKYLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHEL 84
Y A + +Y KYL K+ R G WV+E L P Y MFRME H+ KL L
Sbjct: 166 YGAAMIAYDYHMKYLMKQGNRFLYSHGWNWVRETL-NTPGESYNMFRMETHVVLKLEKLL 342
Query: 85 VEHD-LKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETV 128
V L S M E + +FL + NR IQ +F SGETV
Sbjct: 343 VSKGWLHPSNEMTFLEALIIFLWSCAYSETNRNIQNKFGKSGETV 477
>BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 407
Score = 54.3 bits (129), Expect(2) = 3e-08
Identities = 26/39 (66%), Positives = 29/39 (73%)
Frame = +2
Query: 243 EVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQ 281
E+FNY HSSLR TIER FGV KNRF IL+ MP +K Q
Sbjct: 140 ELFNYRHSSLRMTIERCFGVLKNRFPILKLMPSYKPSRQ 256
Score = 20.8 bits (42), Expect(2) = 3e-08
Identities = 10/30 (33%), Positives = 17/30 (56%)
Frame = +3
Query: 285 VVATMAIHNFIRRSAEMDVDFNLYEDENTV 314
V A AIHN+I + D F ++E+ + +
Sbjct: 264 VTACCAIHNYICKWNLPDELFRIWEEMDDI 353
>BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.40 -
Arabidopsis thaliana, partial (11%)
Length = 788
Score = 43.5 bits (101), Expect = 1e-04
Identities = 35/118 (29%), Positives = 61/118 (51%), Gaps = 7/118 (5%)
Frame = +2
Query: 241 HNEVFNYYHSSLRCTIER---TFGV----WKNRFAILRSMPKFKYETQVHIVVATMAIHN 293
H+ +F YY+++ ++E +F V +K+ F I +S P F Y+T+ IV+A + +HN
Sbjct: 428 HHAIF-YYNTNFHFSVEYVIVSFLV*VCRFKSLFLIFKSAPPFPYKTKKEIVLACVGLHN 604
Query: 294 FIRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
F+R+ D +F +D V +D + N N + V + +HA N +R I
Sbjct: 605 FLRKFCRTD-NFPEEKDSEDV----EDVKEVN-NNDE*ILVTQDQQREHA-NQVRATI 757
>AL374958 GP|8979817|emb| dJ447E21.2 (Similar to X.laevis Xrel2 protein)
{Homo sapiens}, partial (73%)
Length = 524
Score = 34.3 bits (77), Expect = 0.076
Identities = 18/38 (47%), Positives = 22/38 (57%)
Frame = +3
Query: 269 ILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFN 306
IL M F Q I+VAT+A+HNFIR D +FN
Sbjct: 15 ILCDMRPFPLIKQEKIIVATIALHNFIRICGVQDEEFN 128
>TC86370 similar to GP|15451010|gb|AAK96776.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1050
Score = 32.7 bits (73), Expect = 0.22
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 14/95 (14%)
Frame = -3
Query: 234 RSSGFENHNEV----------FNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVH 283
RSS F N + FN++ S L C RT + ++L +P K E
Sbjct: 715 RSSFFSTFNSLIFS*RPELASFNFWASPLNCVTIRTASIRMAALSVLAGLPGSKMEPNSI 536
Query: 284 IVVATMAIHNFIRRSAEM----DVDFNLYEDENTV 314
+ ++++ NF R + E+ D D L+ + +++
Sbjct: 535 NLSLSLSLRNFSRHALELVAAHDSDLTLFFEPDSL 431
>TC86369 similar to GP|15451010|gb|AAK96776.1 Unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 1500
Score = 31.2 bits (69), Expect = 0.64
Identities = 23/95 (24%), Positives = 41/95 (42%), Gaps = 14/95 (14%)
Frame = -2
Query: 234 RSSGFENHNEV----------FNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVH 283
RSS F N + FN++ S L C RT + ++L P K E
Sbjct: 926 RSSFFSTFNSLIFS*RPELASFNFWASPLNCVTIRTASIRMAALSVLAGPPGSKMEPNSI 747
Query: 284 IVVATMAIHNFIRRSAEM----DVDFNLYEDENTV 314
+ ++++ NF R + E+ D D L+ + +++
Sbjct: 746 NLSLSLSLRNFSRHALELVAAHDSDLTLFFEPDSL 642
>BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH oxidase
{Arabidopsis thaliana}, partial (17%)
Length = 610
Score = 28.9 bits (63), Expect = 3.2
Identities = 23/59 (38%), Positives = 28/59 (46%), Gaps = 10/59 (16%)
Frame = -1
Query: 193 ANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRC-------ERYHLP---EFRRSSGFENH 241
A+L F P Q KY+LVD +P + G YR +YHLP FR SS H
Sbjct: 340 AHLTFSLPKQLKYFLVDK--VSPPSHDGEYRSI*QGKVKVQYHLPLI*GFRTSSDMVLH 170
>TC87056 weakly similar to GP|20521237|dbj|BAB91753. contains ESTs
C73990(E30106) AU029295(E30106)~unknown protein, partial
(70%)
Length = 1381
Score = 27.7 bits (60), Expect = 7.1
Identities = 19/59 (32%), Positives = 30/59 (50%)
Frame = -3
Query: 82 HELVEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVRVIDGTHVSCVV 140
H +V+H SKH+ EE A++ V +G+R+I ++ R I T V C+V
Sbjct: 1091 HPIVQHISSQSKHVRSEE--AIYRAVCNQIIGSRLI*KQEC**SRGTRRIYFTQVFCIV 921
>BQ144266 similar to GP|19887436|gb| Fe-S oxidoreductase family protein
{Methanopyrus kandleri AV19}, partial (2%)
Length = 1120
Score = 27.7 bits (60), Expect = 7.1
Identities = 14/50 (28%), Positives = 22/50 (44%)
Frame = +2
Query: 162 VCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSG 211
VC + F+ +L+G + T H R+F K + PP + Y G
Sbjct: 176 VCSPPLSFSVILSGGKNTTHTIRLFHKRAVIKEIK---PPPARSYQARKG 316
>TC93262 weakly similar to PIR|T07972|T07972 leucoanthocyanidin dioxygenase
(EC 1.14.11.-) - common stock, partial (12%)
Length = 681
Score = 27.3 bits (59), Expect = 9.3
Identities = 12/28 (42%), Positives = 15/28 (52%)
Frame = +1
Query: 179 TAHDARVFDKALTTANLNFPHPPQGKYY 206
+ H R + K LT+ NFPH P G Y
Sbjct: 508 SVHFWRDYLKVLTSPQFNFPHKPPGYRY 591
>TC82676 similar to GP|8843849|dbj|BAA97375.1 contains similarity to unknown
protein~gene_id:MWD22.7~pir||T26512 {Arabidopsis
thaliana}, partial (39%)
Length = 849
Score = 27.3 bits (59), Expect = 9.3
Identities = 16/63 (25%), Positives = 28/63 (44%)
Frame = +1
Query: 284 IVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHA 343
I+ +A+ R +D+D + ED N + D +S S+ + +S DH+
Sbjct: 319 IITIQIALKFNCRSILGIDIDSDRVEDANWNLRKTDRLKSARNKPSKVSKLKDNSHTDHS 498
Query: 344 RNS 346
NS
Sbjct: 499 ENS 507
>TC88197 homologue to GP|6739629|gb|AAF27340.1| abscisic acid-activated
protein kinase {Vicia faba}, partial (96%)
Length = 1158
Score = 27.3 bits (59), Expect = 9.3
Identities = 14/52 (26%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Frame = -3
Query: 21 FEESYILAALLGEYATKY-----LCKEPCRTSELTGHAWVQEILQGNPTRCY 67
F + A LLG+Y ++ + E +T + H E+L +PT+C+
Sbjct: 652 FHHTLASAVLLGQYKPEFPQLTLVVNEEPKTWSIQNHRSSNEVLDLHPTKCF 497
>TC78924 similar to GP|20197503|gb|AAC95160.2 putative translation
initiation factor eIF-2B alpha subunit {Arabidopsis
thaliana}, partial (74%)
Length = 981
Score = 27.3 bits (59), Expect = 9.3
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 3/27 (11%)
Frame = -1
Query: 67 YEMFRMEKHI---FHKLCHELVEHDLK 90
YE +R+ +H F LCH L+ HDL+
Sbjct: 906 YETWRIRQHYRFQFCLLCHHLMRHDLR 826
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,221,984
Number of Sequences: 36976
Number of extensions: 214641
Number of successful extensions: 988
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 973
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 983
length of query: 358
length of database: 9,014,727
effective HSP length: 97
effective length of query: 261
effective length of database: 5,428,055
effective search space: 1416722355
effective search space used: 1416722355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC127428.4


