

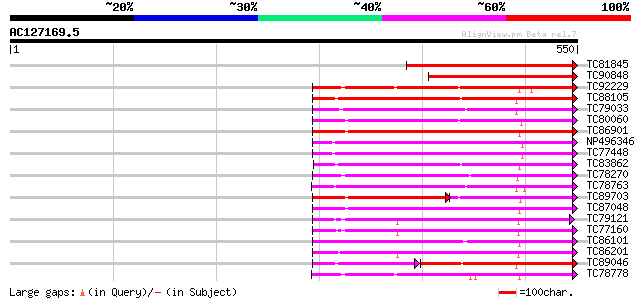
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.5 + phase: 0 /pseudo
(550 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81845 similar to PIR|T01617|T01617 probable protein kinase [im... 249 3e-66
TC90848 similar to GP|15810235|gb|AAL07235.1 putative Pto kinase... 209 3e-54
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 208 4e-54
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 208 5e-54
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 198 5e-51
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 198 5e-51
TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120... 196 1e-50
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 193 2e-49
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 193 2e-49
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 187 7e-48
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 186 2e-47
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 186 2e-47
TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serin... 121 3e-47
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 184 1e-46
TC79121 similar to GP|9651971|gb|AAF91337.1| Pti1 kinase-like pr... 182 2e-46
TC77160 similar to PIR|T48014|T48014 serine/threonine protein ki... 181 5e-46
TC86101 similar to GP|15217283|gb|AAK92627.1 Putative leucine-ri... 181 8e-46
TC86201 similar to PIR|T49003|T49003 protein kinase-like protein... 180 1e-45
TC89046 weakly similar to GP|21740816|emb|CAD41006. OSJNBa0042L1... 132 5e-45
TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-spe... 175 4e-44
>TC81845 similar to PIR|T01617|T01617 probable protein kinase [imported] -
Arabidopsis thaliana, partial (42%)
Length = 762
Score = 249 bits (635), Expect = 3e-66
Identities = 108/165 (65%), Positives = 138/165 (83%)
Frame = +2
Query: 386 RYKIAIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHA 445
R+KI +G ARGLHYLHK CK RIIHRDIKASN+LL D+EPQI+DFGLAKWLP++WTHH+
Sbjct: 8 RHKIVVGTARGLHYLHKGCKRRIIHRDIKASNILLTKDFEPQISDFGLAKWLPSQWTHHS 187
Query: 446 VIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDSSKQNILLWAKPLME 505
+ P+EGTFG+LAPE ++HG+VDEKTD+FAFGV LLE+++GR+PVD S Q++ WAKP++
Sbjct: 188 IAPIEGTFGHLAPEYYLHGVVDEKTDVFAFGVFLLEVISGRKPVDVSHQSLHSWAKPILN 367
Query: 506 SGNIAELADPRMEGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
G+I EL D R+EG YDV QL R+ AS C+R ++ WRP+MTEV
Sbjct: 368 KGDIEELVDARLEGEYDVTQLKRLAFAASLCIRASSTWRPSMTEV 502
>TC90848 similar to GP|15810235|gb|AAL07235.1 putative Pto kinase interactor
{Arabidopsis thaliana}, partial (21%)
Length = 919
Score = 209 bits (531), Expect = 3e-54
Identities = 96/145 (66%), Positives = 120/145 (82%), Gaps = 1/145 (0%)
Frame = +3
Query: 407 RIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIV 466
RIIHRDIKASN+LL +YE +I+DFGLAKWLPN W HH V P+EGTFGYLAPE FMHG+V
Sbjct: 6 RIIHRDIKASNILLNDNYEAEISDFGLAKWLPNNWAHHVVFPIEGTFGYLAPEYFMHGVV 185
Query: 467 DEKTDIFAFGVLLLEIVTGRRPVDS-SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQ 525
DEKTD+FAFGVLLLE++TGRR VDS SKQ++++WAKPL++S N+ ELADPR+E +YD +
Sbjct: 186 DEKTDVFAFGVLLLELITGRRAVDSDSKQSLVIWAKPLLDSKNVQELADPRLEEKYDPTE 365
Query: 526 LYRVVLTASYCVRQTAIWRPAMTEV 550
+ R + TAS CV ++ RP M +V
Sbjct: 366 MNRAMKTASLCVHHSSSKRPFMKQV 440
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 208 bits (530), Expect = 4e-54
Identities = 115/266 (43%), Positives = 167/266 (62%), Gaps = 9/266 (3%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF-E 352
GG+ VYK + +G A+K L + E+EF E+ I V H + SL+GYC E
Sbjct: 221 GGFGRVYKALMPDGRVGALKLLKAGSGQG--EREFRAEVDTISRVHHRHLVSLIGYCIAE 394
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
LI+ + NGNL LH N LDWP R KIAIG ARGL YLH+ C +IIHRD
Sbjct: 395 QQRVLIYEFVPNGNLDQHLHE--SQWNVLDWPKRMKIAIGAARGLAYLHEGCNPKIIHRD 568
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+LL YE Q+ DFGLA+ L + H V GTFGY+APE G + +++D+
Sbjct: 569 IKSSNILLDDSYEAQVADFGLAR-LTDDTNTHVSTRVMGTFGYMAPEYATSGKLTDRSDV 745
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM----ESGNIAELADPRMEGRYDVE 524
F+FGV+LLE+VTGR+PVD ++ ++++ WA+P++ E+G+ +ELADPR+ +Y
Sbjct: 746 FSFGVVLLELVTGRKPVDPTQPVGDESLVEWARPILLRAIETGDFSELADPRLHRQYIDS 925
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
+++R++ A+ C+R +A RP M ++
Sbjct: 926 EMFRMIEAAAACIRHSAPKRPRMVQI 1003
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 208 bits (529), Expect = 5e-54
Identities = 114/262 (43%), Positives = 160/262 (60%), Gaps = 5/262 (1%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGY VY+G L NG +A+K+L N EKEF +E+ IGHV H N LLG+C E
Sbjct: 752 GGYGVVYQGQLINGNPVAIKKLL--NNLGQAEKEFRVEVEAIGHVRHKNLVRLLGFCIEG 925
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
LI+ Y NGNL LHG + L W R KI +G A+ L YLH+ + +++HRD
Sbjct: 926 THRLLIYEYVNNGNLEQWLHGAMRQYGYLTWDARIKILLGTAKALAYLHEAIEPKVVHRD 1105
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+L+ D+ +I+DFGLAK L H V GTFGY+APE G+++EK+D+
Sbjct: 1106 IKSSNILIDDDFNAKISDFGLAKLL-GAGKSHITTRVMGTFGYVAPEYANSGLLNEKSDV 1282
Query: 473 FAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGVLLLE +TGR PVD +++ N++ W K ++ + + E+ DP +E R L R
Sbjct: 1283 YSFGVLLLEAITGRDPVDYNRSAAEVNLVDWLKMMVGNRHAEEVVDPNIETRPSTSALKR 1462
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
V+LTA CV + RP M++V
Sbjct: 1463 VLLTALRCVDPDSEKRPKMSQV 1528
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 198 bits (503), Expect = 5e-51
Identities = 118/263 (44%), Positives = 155/263 (58%), Gaps = 6/263 (2%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAK-DNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
GG+ VY G L +G IAVKRL NK + EF +E+ I+ V H N SL GYC E
Sbjct: 490 GGFGSVYWGQLWDGSQIAVKRLKVWSNK---ADMEFAVEVEILARVRHKNLLSLRGYCAE 660
Query: 353 NGLYLI-FNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
LI ++Y N +L + LHG + LDW R IAIG A G+ YLH IIHR
Sbjct: 661 GQERLIVYDYMPNLSLLSHLHGQHSTESLLDWNRRMNIAIGSAEGIVYLHVQATPHIIHR 840
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+KASNVLL D++ ++ DFG AK +P+ T H V+GT GYLAPE M G +E D
Sbjct: 841 DVKASNVLLDSDFQARVADFGFAKLIPDGAT-HVTTRVKGTLGYLAPEYAMLGKANESCD 1017
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
+++FG+LLLE+ +G++P++ S K+ I WA PL +ELADPR+ G Y E+L
Sbjct: 1018VYSFGILLLELASGKKPLEKLSSSVKRAINDWALPLACEKKFSELADPRLNGDYVEEELK 1197
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
RV+L A C + RP M EV
Sbjct: 1198RVILVALICAQNQPEKRPTMVEV 1266
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 198 bits (503), Expect = 5e-51
Identities = 109/262 (41%), Positives = 157/262 (59%), Gaps = 5/262 (1%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
GG+ VYKG L +G IAVK+L+ +K N+E F+ E+G+I + HPN L G C E
Sbjct: 220 GGFGPVYKGVLSDGAVIAVKQLSSKSKQGNRE--FVNEIGMISALQHPNLVKLYGCCIEG 393
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
N L L++ Y +N +L+ AL G + +LDW R KI +G+ARGL YLH+ + +I+HRD
Sbjct: 394 NQLLLVYEYMENNSLARALFGKPEQRLNLDWRTRMKICVGIARGLAYLHEESRLKIVHRD 573
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+NVLL + +I+DFGLAK L + H + GT GY+APE M G + +K D+
Sbjct: 574 IKATNVLLDKNLNAKISDFGLAK-LDEEENTHISTRIAGTIGYMAPEYAMRGYLTDKADV 750
Query: 473 FAFGVLLLEIVTGRRPVDSSKQN----ILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGV+ LEIV+G + + +L WA L E GN+ EL DP + +Y E+ R
Sbjct: 751 YSFGVVALEIVSGMSNTNYRPKEEFVYLLDWAYVLQEQGNLLELVDPTLGSKYSSEEAMR 930
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
++ A C + RP M+ V
Sbjct: 931 MLQLALLCTNPSPTLRPPMSSV 996
>TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120
{Arabidopsis thaliana}, partial (83%)
Length = 2028
Score = 196 bits (499), Expect = 1e-50
Identities = 106/264 (40%), Positives = 161/264 (60%), Gaps = 7/264 (2%)
Frame = +1
Query: 294 GGYSEVYKGDL-CNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
GG+ VYKG L G+ +AVK+L ++ N+E FL+E+ ++ + PN SL+GYC +
Sbjct: 556 GGFGRVYKGRLETTGQAVAVKQLDRNGLQGNRE--FLVEVLMLSLLHSPNLVSLIGYCAD 729
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ + G+L LH LDW R KIA G A+GL YLH +I+R
Sbjct: 730 GDQRLLVYEFMPLGSLEDHLHDLPADKEPLDWNTRMKIAAGAAKGLEYLHDKANPPVIYR 909
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D K+SN+LL Y P+++DFGLAK P H V GT+GY APE M G + K+D
Sbjct: 910 DFKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSD 1089
Query: 472 IFAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM-ESGNIAELADPRMEGRYDVEQL 526
+++FGV+ LE++TGR+ +DS++ QN++ WA+PL + ++LADPR++GRY + L
Sbjct: 1090VYSFGVVFLELITGRKAIDSTRPHGEQNLVTWARPLFNDRRKFSKLADPRLQGRYPMRGL 1269
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
Y+ + AS C+++ A RP + +V
Sbjct: 1270YQALAVASMCIQEQAAARPLIGDV 1341
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 193 bits (490), Expect = 2e-49
Identities = 100/262 (38%), Positives = 152/262 (57%), Gaps = 5/262 (1%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
GG+ VY+G L +G+ +AVK + + +EF EL ++ + H N LLGYC E
Sbjct: 1741 GGFGSVYRGTLDDGQEVAVK--VRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEY 1914
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+ L++ + NG+L L+G LDWP R IA+G ARGL YLH +IHRD
Sbjct: 1915 DQQILVYPFMSNGSLLDRLYGEASKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRD 2094
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SN+LL ++ DFG +K+ P + + + V GT GYL PE + + EK+D+
Sbjct: 2095 VKSSNILLDQSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDV 2274
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNI----LLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
F+FGV+LLEIV+GR P++ + I + WAKP + + + E+ DP ++G Y E L+R
Sbjct: 2275 FSFGVVLLEIVSGREPLNIKRPRIEWSLVEWAKPYIRASKVDEIVDPGIKGGYHAEALWR 2454
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
VV A C+ + +RP M ++
Sbjct: 2455 VVEVALQCLEPYSTYRPCMVDI 2520
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 193 bits (490), Expect = 2e-49
Identities = 100/262 (38%), Positives = 152/262 (57%), Gaps = 5/262 (1%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
GG+ VY+G L +G+ +AVK + + +EF EL ++ + H N LLGYC E
Sbjct: 2185 GGFGSVYRGTLDDGQEVAVK--VRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEY 2358
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+ L++ + NG+L L+G LDWP R IA+G ARGL YLH +IHRD
Sbjct: 2359 DQQILVYPFMSNGSLLDRLYGEASKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRD 2538
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SN+LL ++ DFG +K+ P + + + V GT GYL PE + + EK+D+
Sbjct: 2539 VKSSNILLDQSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDV 2718
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNI----LLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
F+FGV+LLEIV+GR P++ + I + WAKP + + + E+ DP ++G Y E L+R
Sbjct: 2719 FSFGVVLLEIVSGREPLNIKRPRIEWSLVEWAKPYIRASKVDEIVDPGIKGGYHAEALWR 2898
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
VV A C+ + +RP M ++
Sbjct: 2899 VVEVALQCLEPYSTYRPCMVDI 2964
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 187 bits (476), Expect = 7e-48
Identities = 105/261 (40%), Positives = 153/261 (58%), Gaps = 5/261 (1%)
Frame = +1
Query: 295 GYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFENG 354
GY VY+G L +G +AVK L N EKEF +E+ IG V H N L+GYC E
Sbjct: 79 GYGIVYRGILQDGSIVAVKNLL--NNKGQAEKEFKVEVEAIGKVRHKNLVGLVGYCAEGA 252
Query: 355 L-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
L++ Y NGNL LHG + L W IR KIA+G A+GL YLH+ + +++HRD+
Sbjct: 253 KRMLVYEYVDNGNLEQWLHGDVGPVSPLTWDIRMKIAVGTAKGLAYLHEGLEPKVVHRDV 432
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K+SN+LL + +++DFGLAK L + V GTFGY++PE G+++E +D++
Sbjct: 433 KSSNILLDKKWHAKVSDFGLAKLL-GSGKSYVTTRVMGTFGYVSPEYASTGMLNEGSDVY 609
Query: 474 AFGVLLLEIVTGRRPVDSSK----QNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRV 529
+FG+LL+E+VTGR P+D S+ N++ W K ++ S EL DP +E + L R
Sbjct: 610 SFGILLMELVTGRSPIDYSRAPAEMNLVDWFKGMVASRRGEELVDPLIEIQPSPRSLKRA 789
Query: 530 VLTASYCVRQTAIWRPAMTEV 550
+L C+ A RP M ++
Sbjct: 790 LLVCLRCIDLDANKRPKMGQI 852
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 186 bits (473), Expect = 2e-47
Identities = 108/262 (41%), Positives = 152/262 (57%), Gaps = 5/262 (1%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VYKG L G+ A+K L+ ++K KEFL E+ +I + H N L G C E
Sbjct: 336 GGFGSVYKGVLKGGKLAAIKVLSTESKQG--VKEFLTEINVISEIKHENLVILYGCCVEG 509
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++NY +N +LS L G + DW R +I +GVARGL +LH+ I+HRD
Sbjct: 510 DHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRRRICLGVARGLAFLHEEVLPHIVHRD 689
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL D P+I+DFGLAK +P+ TH + V GT GYLAPE + G + K DI
Sbjct: 690 IKASNILLDKDLTPKISDFGLAKLIPSYMTHVST-RVAGTIGYLAPEYAIRGQLTRKADI 866
Query: 473 FAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGVLL+EIV+GR ++ + Q IL L E +A+L D + G +D E+ +
Sbjct: 867 YSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYERKELAQLVDISLNGEFDAEEACK 1046
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
++ A C + T RP M+ V
Sbjct: 1047ILKIALLCTQDTPKLRPTMSSV 1112
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 186 bits (473), Expect = 2e-47
Identities = 101/265 (38%), Positives = 156/265 (58%), Gaps = 7/265 (2%)
Frame = +3
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGG+ +VYKG L +G +AVKRL K+ + P E +F E+ +I H N L G+C
Sbjct: 1356 RGGFGKVYKGRLADGSLVAVKRL-KEERTPGGELQFQTEVEMISMAVHRNLLRLRGFCMT 1532
Query: 353 -NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ Y NG++++ L LDWP R +IA+G ARGL YLH C +IIHR
Sbjct: 1533 PTERLLVYPYMANGSVASCLRERPPHQEPLDWPTRKRIALGSARGLSYLHDHCDPKIIHR 1712
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+KA+N+LL ++E + DFGLAK + K T H V GT G++APE G EKTD
Sbjct: 1713 DVKAANILLDEEFEAVVGDFGLAKLMDYKDT-HVTTAVRGTIGHIAPEYLSTGKSSEKTD 1889
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILL--WAKPLMESGNIAELADPRMEGRYDVEQ 525
+F +G++LLE++TG+R D ++ +++L W K L++ + L DP ++ Y +
Sbjct: 1890 VFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEMLVDPDLKTNYIEAE 2069
Query: 526 LYRVVLTASYCVRQTAIWRPAMTEV 550
+ +++ A C + + + RP M++V
Sbjct: 2070 VEQLIQVALLCTQGSPMDRPKMSDV 2144
>TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serine/threonine
kinase {Arabidopsis thaliana}, partial (26%)
Length = 1212
Score = 121 bits (304), Expect(2) = 3e-47
Identities = 64/135 (47%), Positives = 86/135 (63%), Gaps = 1/135 (0%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VYKG L +G IAVK L+ +K N+E FL E+G+I + HP L G C E
Sbjct: 59 GGFGPVYKGRLSDGTLIAVKLLSSKSKQGNRE--FLNEIGMISALQHPQLVKLYGCCVEG 232
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L LI+ Y +N +L+ AL G + LDWP RYKI +G+ARGL YLH+ + +++HRD
Sbjct: 233 DQLMLIYEYLENNSLARALFGPAEHQIRLDWPTRYKICVGIARGLAYLHEESRLKVVHRD 412
Query: 413 IKASNVLLGPDYEPQ 427
IKA+NVLL D P+
Sbjct: 413 IKATNVLLDKDLNPK 457
Score = 85.5 bits (210), Expect(2) = 3e-47
Identities = 47/128 (36%), Positives = 73/128 (56%), Gaps = 4/128 (3%)
Frame = +3
Query: 427 QITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGR 486
+I+DFGLAK L + H + GT+GY+APE MHG + +K D+++FG++ LEI+ G
Sbjct: 456 KISDFGLAK-LDEEENTHISTRIAGTYGYMAPEYAMHGYLTDKADVYSFGIVALEILHGS 632
Query: 487 RPVDSSKQ----NILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTASYCVRQTAI 542
++ ++L WA L E GN EL D R+ ++ E+ ++ A C T+
Sbjct: 633 NNTILRQKEEAFHLLDWAHILKEKGNEIELVDKRLGSNFNKEEAMLMINVALLCTNVTSS 812
Query: 543 WRPAMTEV 550
RPAM+ V
Sbjct: 813 LRPAMSSV 836
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 184 bits (466), Expect = 1e-46
Identities = 97/263 (36%), Positives = 157/263 (58%), Gaps = 6/263 (2%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ +V+KG L GE +AVK+L+ D + +E F+ E+ ++ + H N L+GYC +
Sbjct: 493 GGFGKVFKGRLSTGELVAVKQLSHDGRQGFQE--FVTEVLMLSLLHHSNLVKLIGYCTDG 666
Query: 354 GL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y G+L L + L W R KIA+G ARGL YLH +I+RD
Sbjct: 667 DQRLLVYEYMPMGSLEDHLFDLPQDKEPLSWSSRMKIAVGAARGLEYLHCKADPPVIYRD 846
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K++N+LL D+ P+++DFGLAK P H V GT+GY APE M G + K+DI
Sbjct: 847 LKSANILLDSDFSPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDI 1026
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKP-LMESGNIAELADPRMEGRYDVEQLY 527
++FGV+LLE++TGRR +D+SK QN++ W++P + +ADP ++G + V L+
Sbjct: 1027YSFGVVLLELITGRRAIDASKKPGEQNLVSWSRPYFSDRRKFVHMADPLLQGHFPVRCLH 1206
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
+ + + C+++ +RP + ++
Sbjct: 1207QAIAITAMCLQEQPKFRPLIGDI 1275
>TC79121 similar to GP|9651971|gb|AAF91337.1| Pti1 kinase-like protein
{Glycine max}, partial (96%)
Length = 1283
Score = 182 bits (463), Expect = 2e-46
Identities = 100/265 (37%), Positives = 154/265 (57%), Gaps = 10/265 (3%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
G Y +VY+ L NG +A+K+L +K P ++EFL ++ I+ + H N L+ YC +
Sbjct: 372 GAYGKVYRATLKNGREVAIKKL-DSSKQP--DQEFLSQVSIVSRLKHENVVELVNYCVDG 542
Query: 354 GLY-LIFNYSQNGNLSTALHGY-----GKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHR 407
L L + Y+ NG+L LHG + G L W R KIA+G ARGL YLH+ +
Sbjct: 543 PLRALAYEYAPNGSLHDILHGRKGVKGAEPGQVLSWAERVKIAVGAARGLEYLHEKAEVH 722
Query: 408 IIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVD 467
I+HR IK+SN+LL D +I DF L+ P+ V GTFGY APE M G +
Sbjct: 723 IVHRYIKSSNILLFEDGVAKIADFDLSNQAPDAAARLHSTRVLGTFGYHAPEYAMTGNLS 902
Query: 468 EKTDIFAFGVLLLEIVTGRRPVDSS----KQNILLWAKPLMESGNIAELADPRMEGRYDV 523
K+D+++FGV+LLE++TGR+PVD + +Q+++ WA P + + + D R++G Y
Sbjct: 903 SKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVKQCVDVRLKGEYPS 1082
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMT 548
+ + ++ A+ CV+ A +RP M+
Sbjct: 1083KSVAKLAAVAALCVQYEAEFRPNMS 1157
>TC77160 similar to PIR|T48014|T48014 serine/threonine protein kinase-like
protein - Arabidopsis thaliana, partial (94%)
Length = 1493
Score = 181 bits (460), Expect = 5e-46
Identities = 103/267 (38%), Positives = 155/267 (57%), Gaps = 10/267 (3%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
G Y VY G L +G+ A+K+L +K P ++EFL ++ ++ + H N LLGYC +
Sbjct: 429 GSYGRVYYGVLKSGQAAAIKKLDA-SKQP--DEEFLAQVSMVSRLKHDNFVQLLGYCVDG 599
Query: 353 NGLYLIFNYSQNGNLSTALHGY-----GKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHR 407
N L + ++ NG+L LHG + G L W R KIA+G ARGL YLH+
Sbjct: 600 NSRILAYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPH 779
Query: 408 IIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVD 467
IIHRDIK+SNVL+ D +I DF L+ P+ V GTFGY APE M G ++
Sbjct: 780 IIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLN 959
Query: 468 EKTDIFAFGVLLLEIVTGRRPVDSS----KQNILLWAKPLMESGNIAELADPRMEGRYDV 523
K+D+++FGV+LLE++TGR+PVD + +Q+++ WA P + + + D R+ G Y
Sbjct: 960 AKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPP 1139
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ + ++ A+ CV+ A +RP M+ V
Sbjct: 1140KAVAKMAAVAALCVQYEADFRPNMSIV 1220
>TC86101 similar to GP|15217283|gb|AAK92627.1 Putative leucine-rich repeat
transmembrane protein kinase 1 {Oryza sativa}, partial
(64%)
Length = 2629
Score = 181 bits (458), Expect = 8e-46
Identities = 95/263 (36%), Positives = 155/263 (58%), Gaps = 6/263 (2%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
G + VY+ +G+ +AVK++ N +F+ + + + HPN L+GYC E+
Sbjct: 1539 GSFGRVYRAQFDDGQVLAVKKIDSSVLPNNLSDDFMEIVSNLSRLHHPNVTELIGYCSEH 1718
Query: 354 GLYL-IFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G +L ++ Y +NG+L LH L W R K+A+G+AR L YLH+ C ++H++
Sbjct: 1719 GQHLLVYEYHKNGSLHDFLHLPDDYIKPLIWNSRVKVALGIARALEYLHEICSPSVVHKN 1898
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+N+LL D P ++D GLA ++PN + V+ GY APEV + G K+D+
Sbjct: 1899 IKAANILLDADLNPHLSDSGLASYIPNT---NQVLNNNSGSGYDAPEVGLTGQYTLKSDV 2069
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKP-LMESGNIAELADPRMEGRYDVEQLY 527
++FGV++LE+++GR+P DSS+ Q+++ WA P L + +A++ DP +EG Y V+ L
Sbjct: 2070 YSFGVVMLELLSGRKPFDSSRSRFEQSLVRWATPQLHDIDALAKMVDPALEGMYPVKSLS 2249
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
R + CV+ +RP M+EV
Sbjct: 2250 RFADVIALCVQSEPEFRPPMSEV 2318
>TC86201 similar to PIR|T49003|T49003 protein kinase-like protein -
Arabidopsis thaliana, partial (86%)
Length = 2127
Score = 180 bits (457), Expect = 1e-45
Identities = 97/267 (36%), Positives = 156/267 (58%), Gaps = 10/267 (3%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
G Y VY L +G+ +AVK+L +P EFL ++ ++ + + N L GYC E
Sbjct: 441 GSYGRVYYATLNDGKAVAVKKLDVST-EPESNNEFLTQVSMVSRLKNENFVELHGYCVEG 617
Query: 354 GL-YLIFNYSQNGNLSTALHGY-----GKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHR 407
L L + ++ G+L LHG + G +L+W R +IA+ ARGL YLH+ +
Sbjct: 618 NLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLNWMQRVRIAVDAARGLEYLHEKVQPS 797
Query: 408 IIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVD 467
IIHRDI++SNVL+ DY+ ++ DF L+ P+ V GTFGY APE M G +
Sbjct: 798 IIHRDIRSSNVLIFEDYKAKVADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLT 977
Query: 468 EKTDIFAFGVLLLEIVTGRRPVDSS----KQNILLWAKPLMESGNIAELADPRMEGRYDV 523
+K+D+++FGV+LLE++TGR+PVD + +Q+++ WA P + + + DP+++G Y
Sbjct: 978 QKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPP 1157
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ + ++ A+ CV+ A +RP M+ V
Sbjct: 1158KGVAKLAAVAALCVQYEAEFRPNMSIV 1238
>TC89046 weakly similar to GP|21740816|emb|CAD41006. OSJNBa0042L16.4 {Oryza
sativa}, partial (35%)
Length = 1399
Score = 132 bits (333), Expect(2) = 5e-45
Identities = 66/156 (42%), Positives = 98/156 (62%), Gaps = 4/156 (2%)
Frame = +1
Query: 399 YLHKCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAP 458
YLH IIHRDIKASNVLL +++ ++ DFG AK +P H V+GT GYLAP
Sbjct: 574 YLHHEANPHIIHRDIKASNVLLDTEFQAKVADFGFAKLIP-AGVSHLTTRVKGTLGYLAP 750
Query: 459 EVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELAD 514
E M G V E D+++FG+LLLEI++ ++P++ K++I+ W P ++ G +AD
Sbjct: 751 EYAMWGKVSESCDVYSFGILLLEIISAKKPIEKLPGGIKRDIVQWVTPYVQKGVFKHIAD 930
Query: 515 PRMEGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
P+++G +D+EQL V++ A C + RP+M EV
Sbjct: 931 PKLKGNFDLEQLKSVIMIAVRCTDSSPDKRPSMIEV 1038
Score = 67.0 bits (162), Expect(2) = 5e-45
Identities = 40/105 (38%), Positives = 53/105 (50%), Gaps = 1/105 (0%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLG-YCFE 352
GG+ VY G G IAVKRL E EF +E+ ++G V H N L G Y
Sbjct: 258 GGFGSVYWGQTSKGVEIAVKRLK--TMTAKAEMEFAVEVEVLGRVRHKNLLGLRGFYAGG 431
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGL 397
+ ++++Y N +L T LHG + LDWP R I +G A GL
Sbjct: 432 DERLIVYDYMSNHSLLTHLHGQLASDCLLDWPRRMSITVGAAEGL 566
>TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-specific
receptor protein kinase (EC 2.7.1.-) - Arabidopsis
thaliana, partial (23%)
Length = 1808
Score = 175 bits (444), Expect = 4e-44
Identities = 104/273 (38%), Positives = 157/273 (57%), Gaps = 15/273 (5%)
Frame = +3
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGG+ VY G + +G +AVK L+ + KEF E ++ V H N S +GYC E
Sbjct: 750 RGGFGSVYSGQMKDGNKVAVKMLSASSAQG--PKEFQTEAELLMTVHHKNLVSFIGYCDE 923
Query: 353 NG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ LI+ + NGNL L K+ + L W R +IAI A GL YLH CK IIHR
Sbjct: 924 GDKMALIYEFMANGNLKENLSE--KSSHCLSWERRLQIAIDAAEGLDYLHHGCKPPIIHR 1097
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHA---VIPVE---------GTFGYLAPE 459
D+K++N+LL D E +I DFGL+K N +A +I V+ GT GYL PE
Sbjct: 1098 DVKSANILLNEDLEAKIADFGLSKVFKNSDIQNADSTLIHVDVSGEKSAIMGTMGYLDPE 1277
Query: 460 VFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDSSK--QNILLWAKPLMESGNIAELADPRM 517
+ ++EK+DI+++G++LLE++TG V K ++IL + +P + +++++ DPR+
Sbjct: 1278 YYKLQTLNEKSDIYSYGIVLLELITGLPAVIKGKPSKHILEFVRPRLNKDDLSKVIDPRL 1457
Query: 518 EGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
EG++DV ++V+ A C T+I RP M+ V
Sbjct: 1458 EGKFDVSSGWKVLGLAIACTASTSIQRPTMSVV 1556
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.149 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,967,708
Number of Sequences: 36976
Number of extensions: 284282
Number of successful extensions: 3503
Number of sequences better than 10.0: 641
Number of HSP's better than 10.0 without gapping: 2868
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2937
length of query: 550
length of database: 9,014,727
effective HSP length: 101
effective length of query: 449
effective length of database: 5,280,151
effective search space: 2370787799
effective search space used: 2370787799
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC127169.5


