

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.2 - phase: 0
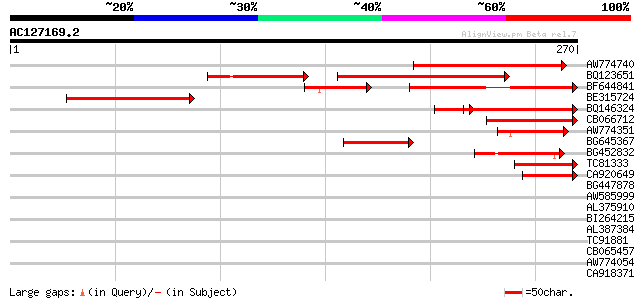
(270 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW774740 130 5e-31
BQ123651 98 1e-30
BF644841 similar to GP|18175662|gb| putative receptor protein ki... 96 5e-28
BE315724 115 1e-26
BQ146324 similar to GP|1945333|emb| ORF YGR160w {Saccharomyces c... 84 6e-20
CB066712 84 6e-17
AW774351 51 4e-07
BG645367 51 5e-07
BG452832 weakly similar to GP|20976571|gb HYPOTHETICAL 90.9 KDA ... 47 8e-06
TC81333 similar to GP|22776289|dbj|BAC12565. ATP-dependent RNA h... 46 1e-05
CA920649 similar to GP|16226812|gb AT5g66680/MSN2_7 {Arabidopsis... 45 3e-05
BG447878 40 0.001
AW585999 36 0.001
AL375910 40 0.001
BI264215 36 0.014
AL387384 similar to GP|6682235|gb|A unknown protein {Arabidopsis... 36 0.014
TC91881 35 0.023
CB065457 28 0.029
AW774054 35 0.040
CA918371 homologue to GP|287405|dbj|BA ORF86 {Oryza sativa (japo... 34 0.052
>AW774740
Length = 739
Score = 130 bits (327), Expect = 5e-31
Identities = 65/73 (89%), Positives = 68/73 (93%)
Frame = +2
Query: 193 RKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIR 252
R+RV VAPIVEKLVENR RWFGHVERRPVDAVVRRV QME+SQVKRGRGRPRKTIRETI
Sbjct: 2 RERVGVAPIVEKLVENRFRWFGHVERRPVDAVVRRVDQMEESQVKRGRGRPRKTIRETI* 181
Query: 253 KDLEVNELDPNLV 265
KD E+NELDPNLV
Sbjct: 182 KDSEINELDPNLV 220
>BQ123651
Length = 654
Score = 97.8 bits (242), Expect(2) = 1e-30
Identities = 46/82 (56%), Positives = 60/82 (73%)
Frame = -2
Query: 157 WAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRVEVAPIVEKLVENRLRWFGHV 216
WA HE ++S A+ RMLRWM TR D+ RND+I RV +APIVEK++E+RLRWFG+V
Sbjct: 248 WAE*CHHERRISEAKKRMLRWMCGNTRRDKIRNDSISVRVGIAPIVEKMIESRLRWFGNV 69
Query: 217 ERRPVDAVVRRVYQMEDSQVKR 238
RP D+VVR+V MED+ + R
Sbjct: 68 WGRPGDSVVRKVDLMEDNPISR 3
Score = 52.8 bits (125), Expect(2) = 1e-30
Identities = 28/48 (58%), Positives = 35/48 (72%)
Frame = -3
Query: 95 YLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFY 142
YLGS +Q + + DV++RIQA +K R VLCDKKVPLK+KGKFY
Sbjct: 421 YLGSVIQK*QK-QRDVNNRIQARLMK*RSPLGVLCDKKVPLKIKGKFY 281
>BF644841 similar to GP|18175662|gb| putative receptor protein kinase
{Arabidopsis thaliana}, partial (1%)
Length = 676
Score = 95.5 bits (236), Expect(2) = 5e-28
Identities = 48/80 (60%), Positives = 59/80 (73%)
Frame = +1
Query: 191 TIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
T +RV V+ IVE +V+N+LRWFG+V RRP+ VVRR G GRPRKT+RET
Sbjct: 298 TTLERVWVSSIVENMVDNKLRWFGYVXRRPIHFVVRR-----------GTGRPRKTVRET 444
Query: 251 IRKDLEVNELDPNLVYDRTL 270
I+KDLE+NELD N+VYDRTL
Sbjct: 445 IKKDLEINELDQNMVYDRTL 504
Score = 46.2 bits (108), Expect(2) = 5e-28
Identities = 21/33 (63%), Positives = 25/33 (75%), Gaps = 1/33 (3%)
Frame = +2
Query: 141 FYRPAF-RPALMYGRECWAVKSQHENQVSVAEM 172
FY F RPA++Y ECWAVK+QHEN+VS EM
Sbjct: 140 FYLLLFLRPAMLYAMECWAVKNQHENKVSYVEM 238
>BE315724
Length = 392
Score = 115 bits (289), Expect = 1e-26
Identities = 57/61 (93%), Positives = 59/61 (96%)
Frame = -2
Query: 28 DVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVEDHII 87
+VVLVGESREEVNGRLETWRQALEAY FRLSRSKTEY+ECNFSGRRSRSTLEVKV DHII
Sbjct: 184 NVVLVGESREEVNGRLETWRQALEAYEFRLSRSKTEYMECNFSGRRSRSTLEVKVGDHII 5
Query: 88 P 88
P
Sbjct: 4 P 2
>BQ146324 similar to GP|1945333|emb| ORF YGR160w {Saccharomyces cerevisiae},
partial (9%)
Length = 705
Score = 84.3 bits (207), Expect(2) = 6e-20
Identities = 40/54 (74%), Positives = 47/54 (86%)
Frame = +3
Query: 217 ERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL 270
ER+ +D VVRRV QMED Q+ RGRGRPRKTI+ TIRKDLE+NELD N++YDRTL
Sbjct: 288 ERKHMDYVVRRVDQMEDGQITRGRGRPRKTIKGTIRKDLEINELD*NMIYDRTL 449
Score = 30.0 bits (66), Expect(2) = 6e-20
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = +1
Query: 203 EKLVENRLRWFGHVERRPV 221
E +VE+RLRWF H+ER +
Sbjct: 244 EYMVESRLRWFEHIERENI 300
>CB066712
Length = 830
Score = 84.0 bits (206), Expect = 6e-17
Identities = 41/43 (95%), Positives = 42/43 (97%)
Frame = +3
Query: 228 VYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL 270
V QME+SQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL
Sbjct: 666 VDQMEESQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL 794
>AW774351
Length = 648
Score = 51.2 bits (121), Expect = 4e-07
Identities = 25/35 (71%), Positives = 29/35 (82%), Gaps = 1/35 (2%)
Frame = +1
Query: 233 DSQVK-RGRGRPRKTIRETIRKDLEVNELDPNLVY 266
+SQ K RGRGRPRKT RETI KDLE+NELD N+ +
Sbjct: 79 NSQTKPRGRGRPRKTTRETIEKDLEINELDKNVFW 183
>BG645367
Length = 739
Score = 50.8 bits (120), Expect = 5e-07
Identities = 20/33 (60%), Positives = 27/33 (81%)
Frame = +1
Query: 160 KSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTI 192
K+QH+N++S +EMRML WM KTRH R RN+T+
Sbjct: 640 KNQHKNKISTSEMRMLHWMCGKTRHGRIRNETL 738
>BG452832 weakly similar to GP|20976571|gb HYPOTHETICAL 90.9 KDA PROTEIN
{Dictyostelium discoideum}, partial (2%)
Length = 669
Score = 47.0 bits (110), Expect = 8e-06
Identities = 25/45 (55%), Positives = 34/45 (75%), Gaps = 2/45 (4%)
Frame = +2
Query: 222 DAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVN--ELDPNL 264
+A++RR+ QM DS + RG+PRKTI +TI++DLEVN LDP L
Sbjct: 101 EALIRRIVQM-DSPIDLDRGQPRKTISQTIKRDLEVNGISLDPIL 232
>TC81333 similar to GP|22776289|dbj|BAC12565. ATP-dependent RNA helicase
{Oceanobacillus iheyensis}, partial (3%)
Length = 756
Score = 46.2 bits (108), Expect = 1e-05
Identities = 22/30 (73%), Positives = 25/30 (83%)
Frame = -1
Query: 241 GRPRKTIRETIRKDLEVNELDPNLVYDRTL 270
GR KTIRETI KDLE+NELD N VY++TL
Sbjct: 756 GRSSKTIRETI*KDLEINELDRNTVYNKTL 667
>CA920649 similar to GP|16226812|gb AT5g66680/MSN2_7 {Arabidopsis thaliana},
partial (38%)
Length = 821
Score = 45.1 bits (105), Expect = 3e-05
Identities = 21/26 (80%), Positives = 24/26 (91%)
Frame = -2
Query: 245 KTIRETIRKDLEVNELDPNLVYDRTL 270
KTI ETIRKDLE+NELD ++VYDRTL
Sbjct: 784 KTI*ETIRKDLEINELDRDMVYDRTL 707
>BG447878
Length = 138
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/26 (65%), Positives = 23/26 (88%)
Frame = +3
Query: 245 KTIRETIRKDLEVNELDPNLVYDRTL 270
KTI ETI+KDLE+NEL+ ++V+DR L
Sbjct: 3 KTITETIKKDLEINELEKDMVFDRPL 80
>AW585999
Length = 446
Score = 36.2 bits (82), Expect(2) = 0.001
Identities = 19/36 (52%), Positives = 24/36 (65%)
Frame = +1
Query: 220 PVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDL 255
PV+ V +V ME S + RGR RKTIRE I+KD+
Sbjct: 40 PVNYAVGKVGXMEGS*ITTCRGRYRKTIREAIKKDI 147
Score = 22.7 bits (47), Expect(2) = 0.001
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +2
Query: 205 LVENRLRWFGHV 216
+VE RLR FGHV
Sbjct: 2 MVETRLRLFGHV 37
>AL375910
Length = 244
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/43 (58%), Positives = 28/43 (64%), Gaps = 8/43 (18%)
Frame = -1
Query: 11 VTEHIQELAPRCM------LFADDVVLVGESREEVNG--RLET 45
+TE+IQELA RCM F DD VL GE REE+ G RLET
Sbjct: 166 LTENIQELALRCMFFFFLFFFTDDAVLHGELREELMGG*RLET 38
>BI264215
Length = 413
Score = 36.2 bits (82), Expect = 0.014
Identities = 18/30 (60%), Positives = 23/30 (76%), Gaps = 4/30 (13%)
Frame = +2
Query: 240 RGRPRKT----IRETIRKDLEVNELDPNLV 265
+GR RKT +RET++KD EVNELD N+V
Sbjct: 86 KGRHRKT*KNLLRETVKKDQEVNELDRNIV 175
>AL387384 similar to GP|6682235|gb|A unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 522
Score = 36.2 bits (82), Expect = 0.014
Identities = 16/20 (80%), Positives = 17/20 (85%)
Frame = +3
Query: 251 IRKDLEVNELDPNLVYDRTL 270
IRKDLEVNE DPN+ YD TL
Sbjct: 255 IRKDLEVNEFDPNMFYD*TL 314
>TC91881
Length = 860
Score = 35.4 bits (80), Expect = 0.023
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Frame = +3
Query: 140 KFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRND--TIRKRVE 197
++Y +++ +CWAVKSQ E S+A+MRML + ++ +N + K+ +
Sbjct: 309 RYYYCTSLKLVIFRIDCWAVKSQ*ERMTSIAKMRMLW*ICGHIGRNKIKNKIRVVNKKKK 488
Query: 198 VAP 200
V P
Sbjct: 489 VYP 497
Score = 34.7 bits (78), Expect = 0.040
Identities = 18/36 (50%), Positives = 22/36 (61%)
Frame = +1
Query: 48 QALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVE 83
Q L F L+RSK EYI C FS + +L+VKVE
Sbjct: 16 QTLNFNDFCLTRSKLEYIGCRFSRGHTNVSLDVKVE 123
>CB065457
Length = 831
Score = 28.5 bits (62), Expect(2) = 0.029
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +3
Query: 110 VSHRIQAGWLKWRRDSD 126
V+H IQ GWLKW+R S+
Sbjct: 48 VNH*IQDGWLKWQRASN 98
Score = 25.4 bits (54), Expect(2) = 0.029
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +1
Query: 94 KYLGSFVQNDGEI 106
KYLGS V NDGEI
Sbjct: 4 KYLGSKVHNDGEI 42
>AW774054
Length = 616
Score = 34.7 bits (78), Expect = 0.040
Identities = 16/20 (80%), Positives = 17/20 (85%)
Frame = +3
Query: 251 IRKDLEVNELDPNLVYDRTL 270
IRKDLEVNE PN+V DRTL
Sbjct: 300 IRKDLEVNEFYPNMVSDRTL 359
>CA918371 homologue to GP|287405|dbj|BA ORF86 {Oryza sativa (japonica
cultivar-group)}, partial (15%)
Length = 596
Score = 34.3 bits (77), Expect = 0.052
Identities = 14/28 (50%), Positives = 22/28 (78%)
Frame = +3
Query: 242 RPRKTIRETIRKDLEVNELDPNLVYDRT 269
RPRK I ++I++DL++N L NL++D T
Sbjct: 87 RPRKNIVQSIKRDLDLNSLSLNLIHDWT 170
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,950,083
Number of Sequences: 36976
Number of extensions: 99864
Number of successful extensions: 483
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 482
length of query: 270
length of database: 9,014,727
effective HSP length: 95
effective length of query: 175
effective length of database: 5,502,007
effective search space: 962851225
effective search space used: 962851225
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC127169.2


