

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.12 + phase: 0
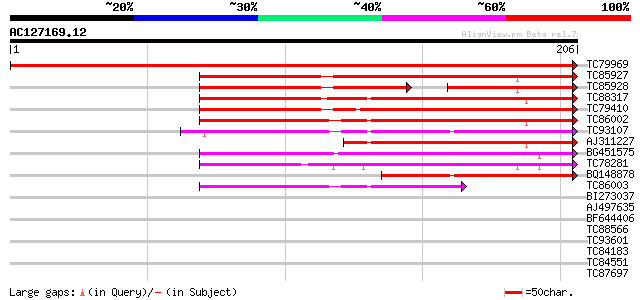
(206 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79969 weakly similar to PIR|T04232|T04232 pathogenesis-related... 449 e-127
TC85927 PR-1 177 3e-45
TC85928 SP|Q40374|PR1_MEDTR Pathogenesis-related protein PR-1 pr... 84 2e-35
TC88317 weakly similar to PIR|T04989|T04989 pathogenesis-related... 141 2e-34
TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-l... 139 1e-33
TC86002 weakly similar to PIR|S10205|S10205 pathogenesis-related... 134 2e-32
TC93107 weakly similar to GP|13560653|gb|AAK30143.1 pathogenesis... 117 2e-27
AJ311227 similar to PIR|H84518|H845 pathogenesis-related PR-1-li... 113 4e-26
BG451575 similar to SP|Q41495|ST14 STS14 protein precursor. [Pot... 90 5e-19
TC78281 weakly similar to GP|14423500|gb|AAK62432.1 Unknown prot... 89 1e-18
BQ148878 weakly similar to GP|4928711|gb|A PR1a precursor {Glyci... 84 4e-17
TC86003 weakly similar to GP|7407641|gb|AAF62171.1| pathogenesis... 79 1e-15
BI273037 similar to GP|7527719|gb|A T5E21.8 {Arabidopsis thalian... 33 0.100
AJ497635 similar to GP|688429|dbj|B tumor-related protein {Nicot... 30 0.65
BF644406 similar to GP|13560653|gb| pathogenesis-related protein... 30 0.65
TC88566 similar to GP|21536824|gb|AAM61156.1 unknown {Arabidopsi... 30 0.85
TC93601 similar to GP|20197913|gb|AAM15308.1 Expressed protein {... 29 1.4
TC84183 similar to GP|9759099|dbj|BAB09668.1 gene_id:MJJ3.15~pir... 28 1.9
TC84551 homologue to GP|14597044|emb|CAC43563. unnamed protein p... 28 1.9
TC87697 similar to PIR|T47802|T47802 hypothetical protein F24G16... 28 2.5
>TC79969 weakly similar to PIR|T04232|T04232 pathogenesis-related protein
homolog F14M19.60 - Arabidopsis thaliana, partial (78%)
Length = 1035
Score = 449 bits (1155), Expect = e-127
Identities = 206/206 (100%), Positives = 206/206 (100%)
Frame = +2
Query: 1 MKSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQL 60
MKSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQL
Sbjct: 53 MKSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQL 232
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 120
CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK
Sbjct: 233 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 412
Query: 121 LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 180
LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR
Sbjct: 413 LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 592
Query: 181 VVCDDGDVFMTCNYDPVGNYVGERPY 206
VVCDDGDVFMTCNYDPVGNYVGERPY
Sbjct: 593 VVCDDGDVFMTCNYDPVGNYVGERPY 670
>TC85927 PR-1
Length = 949
Score = 177 bits (449), Expect = 3e-45
Identities = 79/138 (57%), Positives = 97/138 (70%), Gaps = 1/138 (0%)
Frame = +2
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL N+ RA+ PL+WD +L YA+W+A+QR+ DC +EHS GENI+WGS
Sbjct: 164 QFLIPQNIARAAVGLRPLVWDDKLTHYAQWYANQRRNDCALEHS----NGPYGENIFWGS 331
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGDV 188
G W P AV AW DE+++Y Y NSCV G+MCGHYTQ+VW ST ++GCA VVC DD
Sbjct: 332 GVGWNPAQAVSAWVDEKQFYNYWHNSCVDGEMCGHYTQVVWGSTTKVGCASVVCSDDKGT 511
Query: 189 FMTCNYDPVGNYVGERPY 206
FMTCNYDP GNY GERPY
Sbjct: 512 FMTCNYDPPGNYYGERPY 565
>TC85928 SP|Q40374|PR1_MEDTR Pathogenesis-related protein PR-1 precursor.
[Barrel medic] {Medicago truncatula}, partial (92%)
Length = 795
Score = 83.6 bits (205), Expect(2) = 2e-35
Identities = 35/48 (72%), Positives = 39/48 (80%), Gaps = 1/48 (2%)
Frame = +1
Query: 160 QMCGHYTQIVWKSTRRIGCARVVC-DDGDVFMTCNYDPVGNYVGERPY 206
+MCGHYTQ+VW ST ++GCA VVC DD FMTCNYDP GNY GERPY
Sbjct: 355 EMCGHYTQVVWGSTTKVGCASVVCSDDKGTFMTCNYDPPGNYYGERPY 498
Score = 82.4 bits (202), Expect(2) = 2e-35
Identities = 37/77 (48%), Positives = 49/77 (63%)
Frame = +2
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL N+ RA+ PL+WD +L YA+W+A+QR+ DC +EHS GENI+WGS
Sbjct: 140 QFLIPQNIARAAVGLRPLVWDDKLTHYAQWYANQRRNDCALEHS----NGPYGENIFWGS 307
Query: 130 GSDWTPTDAVKAWADEE 146
G W P AV AW DE+
Sbjct: 308 GVGWNPAQAVSAWVDEK 358
>TC88317 weakly similar to PIR|T04989|T04989 pathogenesis-related protein 1
precursor 19.3K - Arabidopsis thaliana, partial (84%)
Length = 658
Score = 141 bits (355), Expect = 2e-34
Identities = 64/138 (46%), Positives = 96/138 (69%), Gaps = 1/138 (0%)
Frame = +3
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+++ HN R ++WD + +A+ +A+QRK DC++ HS G + GEN+ W S
Sbjct: 120 DYVNAHNDARRQVGVANIVWDNTVASFAQDYANQRKGDCQLIHS--GGGGRYGENLAWSS 293
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-DV 188
G D + +DAVK W +E+ Y Y +N+C SG++CGHYTQ+VW++++R+GCA+V CD+
Sbjct: 294 G-DMSGSDAVKLWVNEKADYDYNSNTCASGKVCGHYTQVVWRNSQRVGCAKVRCDNNRGT 470
Query: 189 FMTCNYDPVGNYVGERPY 206
F+TCNYDP GNYVGE+PY
Sbjct: 471 FITCNYDPPGNYVGEKPY 524
>TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (84%)
Length = 1002
Score = 139 bits (349), Expect = 1e-33
Identities = 62/137 (45%), Positives = 93/137 (67%)
Frame = +2
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+++ HN R ++WD L A+ +A+ R+ DC++ HS G + GEN+ GS
Sbjct: 113 DYVNSHNEARRQVGVANVVWDNNLATVAQNYANSRRGDCRLTHS----GGRYGENLA-GS 277
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVF 189
D + TDAV+ W +E+ Y Y +N+C SG++CGHYTQ+VW++T+RIGCA+V C++G F
Sbjct: 278 TGDLSGTDAVRLWVNEKNDYNYNSNTCASGKVCGHYTQVVWRNTKRIGCAKVRCNNGGTF 457
Query: 190 MTCNYDPVGNYVGERPY 206
+ CNYDP GNYVG++PY
Sbjct: 458 IICNYDPPGNYVGQKPY 508
>TC86002 weakly similar to PIR|S10205|S10205 pathogenesis-related protein 1
- common tobacco, partial (72%)
Length = 830
Score = 134 bits (337), Expect = 2e-32
Identities = 62/138 (44%), Positives = 87/138 (62%), Gaps = 1/138 (0%)
Frame = +1
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
++L HN R+ P+ WD ++ YA + ++ K +CK+ HS GEN+ W S
Sbjct: 130 DYLKIHNKARSDVGVGPISWDAKVASYAETYVNKLKANCKMVHSKGP----YGENLAWSS 297
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-DV 188
G D T T AV W E+KYY Y +NSC G CGHYTQ+VW+ + R+GCA+V C+DG
Sbjct: 298 G-DMTGTAAVTMWIGEKKYYNYNSNSCAVGYQCGHYTQVVWRDSVRVGCAKVKCNDGRST 474
Query: 189 FMTCNYDPVGNYVGERPY 206
++CNYDP GNY+G+RP+
Sbjct: 475 IISCNYDPPGNYIGQRPF 528
>TC93107 weakly similar to GP|13560653|gb|AAK30143.1 pathogenesis-related
protein PR-1 precursor {Capsicum annuum}, partial (68%)
Length = 560
Score = 117 bits (294), Expect = 2e-27
Identities = 64/150 (42%), Positives = 89/150 (58%), Gaps = 6/150 (4%)
Frame = +1
Query: 63 NCMQESL------EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPE 116
+CM SL +FL HN R PL W+ LE YA+ +A++R +C++EHS
Sbjct: 61 SCMNISLAQNSPQDFLEVHNQARDEVGVGPLYWEQTLEAYAQNYANKRIKNCELEHSMGP 240
Query: 117 DGFKLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRI 176
GEN+ G G + TD+VK W E+ Y Y +NSCV+ + CGHYTQI+W+ + +
Sbjct: 241 ----YGENLAEGYG-EVNGTDSVKFWLSEKPNYDYNSNSCVNDE-CGHYTQIIWRDSVHL 402
Query: 177 GCARVVCDDGDVFMTCNYDPVGNYVGERPY 206
GCA+ C +G VF+ C+Y P GN GERPY
Sbjct: 403 GCAKSKCKNGWVFVICSYSPPGNVEGERPY 492
>AJ311227 similar to PIR|H84518|H845 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (52%)
Length = 379
Score = 113 bits (283), Expect = 4e-26
Identities = 48/86 (55%), Positives = 64/86 (73%), Gaps = 1/86 (1%)
Frame = +2
Query: 122 GENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARV 181
GEN+ WGSG D T T AV W E++YY Y +NSC +G CGHYTQ+VW+++ R+GCA+V
Sbjct: 35 GENLAWGSG-DITGTGAVNMWIGEKRYYNYNSNSCAAGYQCGHYTQVVWRNSVRVGCAKV 211
Query: 182 VCDDG-DVFMTCNYDPVGNYVGERPY 206
C++G ++CNYDP GNY G+RPY
Sbjct: 212 KCNNGRSTIISCNYDPPGNYNGQRPY 289
>BG451575 similar to SP|Q41495|ST14 STS14 protein precursor. [Potato]
{Solanum tuberosum}, partial (48%)
Length = 427
Score = 90.1 bits (222), Expect = 5e-19
Identities = 49/138 (35%), Positives = 67/138 (48%), Gaps = 1/138 (0%)
Frame = +1
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
EFL HN RAS PL W QL ++ + + G K G N
Sbjct: 10 EFLQTHNQARASVGVEPLTWSEQLANTTSKLVRYQRDKLSCQFANLTAG-KYGANQLMAR 186
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVF 189
G+ TP V+ W E+++Y + N+CV CG YTQ+VW+ + +GCA+ C D
Sbjct: 187 GAAVTPRMVVEEWVKEKEFYNHSDNTCVVNHRCGVYTQVVWRKSVELGCAQTTCGKEDTX 366
Query: 190 MT-CNYDPVGNYVGERPY 206
++ C Y P GNYVG PY
Sbjct: 367 LSICFYYPPGNYVGXSPY 420
>TC78281 weakly similar to GP|14423500|gb|AAK62432.1 Unknown protein
{Arabidopsis thaliana}, partial (71%)
Length = 760
Score = 89.0 bits (219), Expect = 1e-18
Identities = 51/141 (36%), Positives = 74/141 (52%), Gaps = 4/141 (2%)
Frame = +3
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPE-DGFKLGENIYW- 127
EFL HN RA PL W +L + ++ K+ F K G N W
Sbjct: 183 EFLESHNKARAEVGVEPLQWSEKLAKDTSLLVRYQRN--KMACDFANLTASKYGGNQLWA 356
Query: 128 GSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDG 186
GS + TP+ AV+ W E+++Y +V N+CV CG YTQ+VWK + ++GC++ C
Sbjct: 357 GSAAAVTPSKAVEEWVKEKEFYIHVNNTCVVNHECGVYTQVVWKKSAQLGCSQATCTGKK 536
Query: 187 DVFMT-CNYDPVGNYVGERPY 206
+ +T C YDP GN +GE P+
Sbjct: 537 EASLTICFYDPPGNVIGESPF 599
>BQ148878 weakly similar to GP|4928711|gb|A PR1a precursor {Glycine max},
partial (44%)
Length = 450
Score = 84.0 bits (206), Expect = 4e-17
Identities = 34/71 (47%), Positives = 47/71 (65%)
Frame = +2
Query: 136 TDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVFMTCNYD 195
T AVK WADE+ ++ N C G+ C H+TQ+VW + R+GC +V C++G F+TCNY
Sbjct: 113 TQAVKGWADEKPHFDNYLNKCFDGE-CHHFTQVVWSGSLRLGCGKVKCNNGGTFVTCNYY 289
Query: 196 PVGNYVGERPY 206
P GN G+ PY
Sbjct: 290 PPGNIPGQLPY 322
>TC86003 weakly similar to GP|7407641|gb|AAF62171.1| pathogenesis-related
protein 1 {Betula pendula}, partial (95%)
Length = 356
Score = 79.0 bits (193), Expect = 1e-15
Identities = 40/97 (41%), Positives = 54/97 (55%)
Frame = +1
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
++L HN R+ P+ WD ++ YA + ++ K +CK+ HS GEN+ W S
Sbjct: 58 DYLKIHNKARSDVGVGPISWDAKVASYAETYVNKLKANCKMVHSKGP----YGENLAWSS 225
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYT 166
G D T T AV W E+KYY Y +NSC G CGHYT
Sbjct: 226 G-DMTGTAAVTMWIGEKKYYNYNSNSCAVGYQCGHYT 333
>BI273037 similar to GP|7527719|gb|A T5E21.8 {Arabidopsis thaliana}, partial
(4%)
Length = 392
Score = 32.7 bits (73), Expect = 0.100
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 1/66 (1%)
Frame = +3
Query: 9 CFFIFVT-FTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNCMQE 67
CFFI++ + + S S SSSSSS + +LS N S K K+ WN Q
Sbjct: 48 CFFIYIILWNQRRKSKSNNTSSSSSSINYYFCYFLSSF----NHSSSKEKKESTWNTKQV 215
Query: 68 SLEFLF 73
+ FLF
Sbjct: 216 A*LFLF 233
>AJ497635 similar to GP|688429|dbj|B tumor-related protein {Nicotiana glauca
x Nicotiana langsdorffii}, partial (22%)
Length = 445
Score = 30.0 bits (66), Expect = 0.65
Identities = 18/65 (27%), Positives = 26/65 (39%), Gaps = 25/65 (38%)
Frame = +2
Query: 162 CGHYTQIVWKSTRRIG-CARVVCD------------------------DGDVFMTCNYDP 196
C HYTQ+VW +T++ C R + + G +F C YDP
Sbjct: 5 CRHYTQVVWSNTKQTWLCQRKL*EWMDIFHL*RTTHQEIMSAINLTRIQGLIFFICGYDP 184
Query: 197 VGNYV 201
N+V
Sbjct: 185 A*NHV 199
Score = 27.3 bits (59), Expect = 4.2
Identities = 14/27 (51%), Positives = 17/27 (62%)
Frame = +1
Query: 180 RVVCDDGDVFMTCNYDPVGNYVGERPY 206
+VV DG F Y P GNYVG++PY
Sbjct: 64 KVVRMDGH-FSFVAYYPPGNYVGDKPY 141
>BF644406 similar to GP|13560653|gb| pathogenesis-related protein PR-1
precursor {Capsicum annuum}, partial (25%)
Length = 293
Score = 30.0 bits (66), Expect = 0.65
Identities = 13/47 (27%), Positives = 24/47 (50%)
Frame = +1
Query: 63 NCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCK 109
N +++ HN R ++WD + +A+ +A+QRK DC+
Sbjct: 151 NAQNSRADYVNAHNDARRQVGVGDIVWDNTVASFAQDYANQRKGDCQ 291
>TC88566 similar to GP|21536824|gb|AAM61156.1 unknown {Arabidopsis
thaliana}, partial (65%)
Length = 1432
Score = 29.6 bits (65), Expect = 0.85
Identities = 14/24 (58%), Positives = 16/24 (66%)
Frame = +3
Query: 11 FIFVTFTTKTLSTSPQPSSSSSSP 34
F F T ++L T PQP SSSSSP
Sbjct: 303 FSFKPITFQSLPTKPQPPSSSSSP 374
>TC93601 similar to GP|20197913|gb|AAM15308.1 Expressed protein {Arabidopsis
thaliana}, partial (21%)
Length = 613
Score = 28.9 bits (63), Expect = 1.4
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +1
Query: 14 VTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVS 57
++ +T TLS+ P P SSSS P Q + L Q +K N S ++ S
Sbjct: 97 ISASTNTLSSFPNPHSSSSFPQQ-QQRTLLQNRKIQNLSNHERS 225
>TC84183 similar to GP|9759099|dbj|BAB09668.1
gene_id:MJJ3.15~pir||S77290~similar to unknown protein
{Arabidopsis thaliana}, partial (4%)
Length = 694
Score = 28.5 bits (62), Expect = 1.9
Identities = 10/25 (40%), Positives = 17/25 (68%)
Frame = -3
Query: 6 LLLCFFIFVTFTTKTLSTSPQPSSS 30
+ +CFF+ V T + ++T+ PSSS
Sbjct: 347 IFMCFFLLVVITIRFINTTSNPSSS 273
>TC84551 homologue to GP|14597044|emb|CAC43563. unnamed protein product
{Arabidopsis thaliana}, partial (27%)
Length = 1133
Score = 28.5 bits (62), Expect = 1.9
Identities = 20/56 (35%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Frame = +1
Query: 9 CFFIFV-TFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWN 63
CFFI++ + + S S SSSSSS + +LS N S K K+ WN
Sbjct: 79 CFFIYIILWNQRRKSKSNNTSSSSSSINYYFCYFLSS----FNHSSSKEKKESTWN 234
>TC87697 similar to PIR|T47802|T47802 hypothetical protein F24G16.50 -
Arabidopsis thaliana, partial (7%)
Length = 1514
Score = 28.1 bits (61), Expect = 2.5
Identities = 10/28 (35%), Positives = 17/28 (60%)
Frame = +2
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLM 88
CW M+ S+ L R ++ ++W LPL+
Sbjct: 629 CWESMKHSVTQLIREKMLFEAQWILPLL 712
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.133 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,386,554
Number of Sequences: 36976
Number of extensions: 170689
Number of successful extensions: 1337
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1308
length of query: 206
length of database: 9,014,727
effective HSP length: 92
effective length of query: 114
effective length of database: 5,612,935
effective search space: 639874590
effective search space used: 639874590
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC127169.12


