

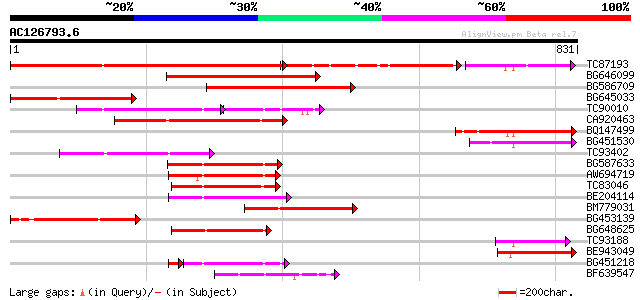
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126793.6 - phase: 0
(831 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Or... 392 e-109
BG646099 weakly similar to GP|13957628|gb putative resistance pr... 355 4e-98
BG586709 weakly similar to GP|2852684|gb|A resistance protein ca... 255 4e-68
BG645033 similar to GP|21616922|gb| NBS-LRR protein {Oryza sativ... 232 4e-61
TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR ty... 171 3e-60
CA920463 weakly similar to GP|2852684|gb|A resistance protein ca... 185 5e-47
BQ147499 177 2e-44
BG451530 homologue to GP|23505092|emb hypothetical protein {Plas... 173 2e-43
TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type put... 172 6e-43
BG587633 similar to GP|20385442|gb| resistance gene analog {Viti... 157 1e-38
AW694719 similar to GP|11994217|db disease resistance comples pr... 150 1e-36
TC83046 weakly similar to GP|10280097|emb|CAC09980. nucleotide s... 148 7e-36
BE204114 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon pi... 139 4e-33
BM779031 weakly similar to GP|6606266|gb| Vrga1 {Aegilops ventri... 137 1e-32
BG453139 similar to GP|5817349|gb| putative NBS-LRR type disease... 137 1e-32
BG648625 weakly similar to GP|20385440|gb| resistance gene analo... 135 5e-32
TC93188 129 4e-30
BE943049 129 6e-30
BG451218 similar to GP|11994217|db disease resistance comples pr... 113 2e-28
BF639547 similar to GP|15553678|gb I2C-5 {Lycopersicon pimpinell... 120 2e-27
>TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 3119
Score = 392 bits (1008), Expect = e-109
Identities = 231/411 (56%), Positives = 278/411 (67%), Gaps = 4/411 (0%)
Frame = +3
Query: 1 MAHEIEKLQTKFNDVVKDMPGLNLNSNVVVVEQSDIVRRETSSFVLESEIIGREDDKKKI 60
MAHEIEK+Q F DV KDM L LNSNVVVV +++ VRRET S+VLESEIIGREDDK KI
Sbjct: 429 MAHEIEKIQRSFKDVEKDMSYLKLNSNVVVVAKTNNVRRETCSYVLESEIIGREDDKNKI 608
Query: 61 ISLLRQSHENQNVSLVAIVGIGGLGKTALAQLVYNDAQVTKSFEKRMWVCVSDNFDVKTI 120
ISLLRQSHE+QNVSLVAIVGIGGLGKTALAQLVY D +V FEK MWVCVSDNFD KTI
Sbjct: 609 ISLLRQSHEHQNVSLVAIVGIGGLGKTALAQLVYKDGEVKNLFEKHMWVCVSDNFDFKTI 788
Query: 121 LKKMLESLTNKKIDDKLSLENLQSMLRDTLTAMRYLLVLDDIWNDSFEKWAQLKTYLMCG 180
LK M+ SLT + + +K +L+ LQSML+ LT RYLLVLDD+WN+SFEKW QL+ YLMCG
Sbjct: 789 LKNMVASLTKEDVVNK-TLQELQSMLQVNLTGKRYLLVLDDVWNESFEKWDQLRPYLMCG 965
Query: 181 AQGSKVVVTTRSKVVAQTMGVSVPYTLNGLTPEKSWSLLKNIVTYGDETKGVLNQTLETI 240
AQGSKVV+TT SK+VA MGVS + L GLTPEKSW L KNIV +GD T GV NQ LE+I
Sbjct: 966 AQGSKVVMTTCSKIVADRMGVSDQHVLRGLTPEKSWVLFKNIV-FGDVTVGV-NQPLESI 1139
Query: 241 GKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCE-EEESIMPVLKLSYHN 299
GKKIA KC GVPLAIR+LGG+L+ +S E+EW+ VLQ + + C E SI+ K+
Sbjct: 1140GKKIAEKCKGVPLAIRSLGGILRSESKESEWINVLQGECLENCVIRENSIIAGPKIELPE 1319
Query: 300 LSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVNIFLMKS 359
L + F I + + + K + E FL +
Sbjct: 1320LITSTKAMFCLLLFISSGLGI*EG*VNSNVDGTRLSWLFS*KAMHERCW*SICKNFLEEF 1499
Query: 360 FLQDVETD-SCGDIHSFKMHDLIHDLAMEVAGND--CCYLDSETKNLVESP 407
L + + S + FKMHDL+HDLA +VAGND ++ + KN+ E P
Sbjct: 1500ILPRCKFE*SWVMVTGFKMHDLMHDLATQVAGNDLLVLWIQVKQKNV*EDP 1652
Score = 193 bits (490), Expect(2) = 6e-89
Identities = 124/265 (46%), Positives = 168/265 (62%), Gaps = 1/265 (0%)
Frame = +2
Query: 398 SETKNLVESPMHIMMKMDDIGLLESVDASRLRTLILMP-NLKTFRNEEDMSIISKFKYLR 456
S+ K + P+H++++ D + LLES+D+SRLRTLI+M N FR E +S+ KYLR
Sbjct: 1622 SKAKKCLGRPVHVLVEYDALCLLESLDSSRLRTLIVMSYNDYKFRRPE-ISVFRNLKYLR 1798
Query: 457 VLKLSHCSLCKLCDSIVKLKHLRYLDLWYCRGVGSVFKSITNMVCLQTLKLVGQKNVPIS 516
LK+ CS + I KLKHLR+LDL S+ KSI N+VCLQT+KL +V S
Sbjct: 1799 FLKMESCSRQGV-GFIEKLKHLRHLDLRNYDSGESLSKSIGNLVCLQTIKL--NDSVVDS 1969
Query: 517 IKDVYNLINLRQLDLDIVMSYEKKNTVCRFGKLCGVGGLYKRLVFSDWHSSLTNLVEISI 576
+ V LINLR L ++ V +K T F KL K L S+W S +TN++E+ +
Sbjct: 1970 PEVVSKLINLRHLTINHVTFKDK--TPSGFRKLSIQQP--KGLSLSNWLSPVTNIIELCL 2137
Query: 577 KKFYTLKYLPPMERLPFLKRLNLFCLDDLEYIYFEEPILPESFFPSLKKLIITDCFKLRG 636
L+YLPP+ERLPFLK L+L L+ LEYIY+EEPIL ESFFPSL+ L +C+KL+G
Sbjct: 2138 YNCQDLRYLPPLERLPFLKSLDLRWLNQLEYIYYEEPILHESFFPSLEILNFFECWKLKG 2317
Query: 637 WWRLRDDVNNVENSSQFHHLSFPPF 661
W R+ DD+N++ +S HHL P F
Sbjct: 2318 WRRMGDDLNDINSS---HHLFLPHF 2383
Score = 154 bits (388), Expect(2) = 6e-89
Identities = 90/192 (46%), Positives = 112/192 (57%), Gaps = 30/192 (15%)
Frame = +1
Query: 668 LSIFSCPMLTCIPTFPNLDKTLHLVSTSVETLEATLNMVGSELAIEFPPLSKLKYLRL-- 725
L I C MLT +PTFPN+ K L L S S E LEATLN+ S+ +I FPPLS LK +
Sbjct: 2398 LVIVRCKMLTFMPTFPNIKKKLELWSCSAEILEATLNIAESQYSIGFPPLSILKSFHIIE 2577
Query: 726 -----------------GGEDLDLKILP---------FFKEDHNFLSSIQNFEFCNCSDL 759
E+LDL L +FK+D L S+Q + C L
Sbjct: 2578 TIMGMENVPKDWLKNLTSLENLDLDSLSSQQFEVIEMWFKDDLICLPSLQKIQIRTCR-L 2754
Query: 760 KVLPDWICNLSSLQHISIQRCRNLASLPEGMPRLSKLHTLEIFGCPL--LVEECVTQTSA 817
K LPDWICN+SSLQH+++ C +L LPEGMPRL+ LHTLEI GCP+ +E + +T A
Sbjct: 2755 KALPDWICNISSLQHLAVYGCESLVDLPEGMPRLTNLHTLEIIGCPISYYYDEFLRKTGA 2934
Query: 818 TWSKISHIPNII 829
TWSKI+HIP II
Sbjct: 2935 TWSKIAHIPKII 2970
>BG646099 weakly similar to GP|13957628|gb putative resistance protein {Oryza
sativa}, partial (4%)
Length = 681
Score = 355 bits (911), Expect = 4e-98
Identities = 170/226 (75%), Positives = 194/226 (85%)
Frame = -3
Query: 230 KGVLNQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESI 289
KGVLNQ LE IGKKIA KCSGVPLAIRTLGGLLQGKS+ETEWVGVLQDDFWKLCE+EE+I
Sbjct: 679 KGVLNQNLEAIGKKIAEKCSGVPLAIRTLGGLLQGKSEETEWVGVLQDDFWKLCEDEETI 500
Query: 290 MPVLKLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGN 349
MPVLKLSY NLSPQLRQCF+YC++YPKDW+I K ELI LWMAQGYLECS +K+ EDIGN
Sbjct: 499 MPVLKLSYQNLSPQLRQCFSYCSLYPKDWEIKKDELIQLWMAQGYLECSTQKQGPEDIGN 320
Query: 350 QFVNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLDSETKNLVESPMH 409
QFV IFLMKSF QD + D CGDI SFKMHDL+HDLAM++AGNDCCY+ SETK L SP+H
Sbjct: 319 QFVKIFLMKSFFQDAKIDDCGDICSFKMHDLMHDLAMQIAGNDCCYIGSETKRLAGSPIH 140
Query: 410 IMMKMDDIGLLESVDASRLRTLILMPNLKTFRNEEDMSIISKFKYL 455
+M+K+D IG LES+DASRLRTLIL+ + +R E +S+I KF L
Sbjct: 139 VMLKLDFIGWLESLDASRLRTLILLHSDLPYRGCEKLSVILKFNRL 2
>BG586709 weakly similar to GP|2852684|gb|A resistance protein candidate
{Lactuca sativa}, partial (6%)
Length = 671
Score = 255 bits (652), Expect = 4e-68
Identities = 126/220 (57%), Positives = 171/220 (77%), Gaps = 1/220 (0%)
Frame = +2
Query: 289 IMPVLKLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIG 348
+M +L L Y +LS +LRQCFAYC+++PKDW+I K LI LWMAQGYLECS K+L+ED+G
Sbjct: 2 LMLILTLGYQSLSLRLRQCFAYCSLFPKDWEIEKDMLIQLWMAQGYLECSDGKQLLEDVG 181
Query: 349 NQFVNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLDSETKNLVESPM 408
N+FV I L+KSF QD + G++ SFKMH+L+HDLA +VAGNDC Y++SE K V+ P+
Sbjct: 182 NEFVKILLIKSFFQDAKEGEDGELVSFKMHNLMHDLATQVAGNDCFYIESEAKRRVQRPV 361
Query: 409 HIMMKMDDIGLLESVDASRLRTLIL-MPNLKTFRNEEDMSIISKFKYLRVLKLSHCSLCK 467
H+ ++ + + LL S+DA+RLRTLIL N + N ++MS+ISKFK LRVL LS+CSL K
Sbjct: 362 HVSLEPNAVHLLGSLDANRLRTLILWSSNEEEELNGDEMSVISKFKCLRVLMLSYCSLSK 541
Query: 468 LCDSIVKLKHLRYLDLWYCRGVGSVFKSITNMVCLQTLKL 507
L +SI KLKHLRYL+L +CRG+GS++KS +++ LQ+L L
Sbjct: 542 LSNSIGKLKHLRYLNLSHCRGLGSLYKSFSSLGLLQSLIL 661
>BG645033 similar to GP|21616922|gb| NBS-LRR protein {Oryza sativa (japonica
cultivar-group)}, partial (20%)
Length = 747
Score = 232 bits (592), Expect = 4e-61
Identities = 119/186 (63%), Positives = 147/186 (78%)
Frame = +1
Query: 1 MAHEIEKLQTKFNDVVKDMPGLNLNSNVVVVEQSDIVRRETSSFVLESEIIGREDDKKKI 60
MA EIEK+Q KF+ V++ M LNL+S V VV+Q+D +R ++ SF+LES I+GREDDKK+I
Sbjct: 175 MAPEIEKVQKKFDVVLEQMSKLNLSSKVPVVKQTDSLRNKSISFLLESNIMGREDDKKEI 354
Query: 61 ISLLRQSHENQNVSLVAIVGIGGLGKTALAQLVYNDAQVTKSFEKRMWVCVSDNFDVKTI 120
I+LL Q H N +S + IVGIGG+GKT LA+ VYND +V K FEK+MWVCVS NFDVKTI
Sbjct: 355 INLLTQPHGN--ISSIVIVGIGGIGKTTLARFVYNDVEVQKHFEKKMWVCVSSNFDVKTI 528
Query: 121 LKKMLESLTNKKIDDKLSLENLQSMLRDTLTAMRYLLVLDDIWNDSFEKWAQLKTYLMCG 180
+KKMLESL ++KIDDKLS E +Q L + LT RYLLVLDDI N S EKW QL+TYLMCG
Sbjct: 529 VKKMLESLIDRKIDDKLSFEYIQQKLHENLTGERYLLVLDDICNASHEKWTQLRTYLMCG 708
Query: 181 AQGSKV 186
A+ S+V
Sbjct: 709 AEDSRV 726
>TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (88%)
Length = 1107
Score = 171 bits (433), Expect(2) = 3e-60
Identities = 87/219 (39%), Positives = 130/219 (58%)
Frame = +3
Query: 98 QVTKSFEKRMWVCVSDNFDVKTILKKMLESLTNKKIDDKLSLENLQSMLRDTLTAMRYLL 157
+V + F+ + WVCVS++FD+ + K +LES+T+ D K +L+ L+ L+ R+L
Sbjct: 12 EVQQHFDLKAWVCVSEDFDIMRVTKSLLESVTSTTWDSK-NLDVLRVELKKISREKRFLF 188
Query: 158 VLDDIWNDSFEKWAQLKTYLMCGAQGSKVVVTTRSKVVAQTMGVSVPYTLNGLTPEKSWS 217
V DD+WND++ W +L + + G GS V++TTR + VA+ + L L+ E WS
Sbjct: 189 VFDDLWNDNYNDWNELASPFINGKPGSMVIITTRQQKVAEVAHTFPIHKLKLLSNEDCWS 368
Query: 218 LLKNIVTYGDETKGVLNQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQD 277
LL DE N LE G+KIA KC G+P+A +T+GGLL+ K D TEW +L
Sbjct: 369 LLSKHALGSDEFHHSSNTALEETGRKIARKCGGLPIAAKTIGGLLRSKVDITEWTSILNS 548
Query: 278 DFWKLCEEEESIMPVLKLSYHNLSPQLRQCFAYCAIYPK 316
+ W L ++I+P L LSY L L++CFAYC+I+ K
Sbjct: 549 NIWNL--RNDNILPALHLSYQYLPSHLKRCFAYCSIFSK 659
Score = 80.1 bits (196), Expect(2) = 3e-60
Identities = 50/158 (31%), Positives = 83/158 (51%), Gaps = 10/158 (6%)
Frame = +1
Query: 314 YPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVNIFLMKSFLQDVETDSCGDIH 373
+PKD+ + + EL+ LWMA+G+L+CS + K ME++G+ L +S +Q + D G+
Sbjct: 652 FPKDYPLDRKELVLLWMAEGFLDCSQRGKKMEELGDDCFAELLSRSLIQQLSDDDRGE-- 825
Query: 374 SFKMHDLIHDLAMEVAGNDCCYLDSETKNLVESPMHIMMKMDDIGLLESVDASR----LR 429
F MHDL++DLA V+G CC L E ++ E+ H ++ + + LR
Sbjct: 826 KFVMHDLVNDLATFVSGKSCCRL--ECGDIPENVRHFSYNQENYDIFMKFEKLHNFKCLR 999
Query: 430 TLILM------PNLKTFRNEEDMSIISKFKYLRVLKLS 461
+ + + N +F+ D ++ K LRVL LS
Sbjct: 1000SFLFICLMTWRDNYLSFKVVND--LLPSHKRLRVLSLS 1107
>CA920463 weakly similar to GP|2852684|gb|A resistance protein candidate
{Lactuca sativa}, partial (6%)
Length = 810
Score = 185 bits (470), Expect = 5e-47
Identities = 102/254 (40%), Positives = 154/254 (60%), Gaps = 1/254 (0%)
Frame = -1
Query: 154 RYLLVLDDIWNDSFEKWAQLKTYLMCGAQGSKVVVTTRSKVVAQTMGVSVPYTLNGLTPE 213
++LL LDD+W++ KW ++K L G +GSKV+VTTRS +A+ M + YTL GL+ E
Sbjct: 753 KFLLRLDDVWSEDRVKWIEVKNLLQVGDEGSKVLVTTRSHSIAKMMCTNTSYTLQGLSRE 574
Query: 214 KSWSL-LKNIVTYGDETKGVLNQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWV 272
S S+ +K G+E K L IGK+I KC G+PLA+RTLG L K D EW
Sbjct: 573 DSLSVFVKWAFKEGEEKK---YPKLIEIGKEIVQKCGGLPLALRTLGSSLFLKDDIEEWK 403
Query: 273 GVLQDDFWKLCEEEESIMPVLKLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQ 332
V ++ W L ++E+ I+P LKLS+ L L++CFA +++ KD+ + + LW A
Sbjct: 402 FVRDNEIWNLPQKEDDILPALKLSFDQLPSYLKRCFACFSLFVKDFHFSNYSVTVLWEAL 223
Query: 333 GYLECSAKKKLMEDIGNQFVNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGND 392
+L K K +ED+GNQF++ +SFLQD G++ FK+HDL+HDLA+ VA ++
Sbjct: 222 DFLPSPNKGKTLEDVGNQFLHELQSRSFLQDFYVS--GNVCVFKLHDLVHDLALYVARDE 49
Query: 393 CCYLDSETKNLVES 406
L +N++++
Sbjct: 48 FQLLKFHNENIIKN 7
>BQ147499
Length = 631
Score = 177 bits (448), Expect = 2e-44
Identities = 103/205 (50%), Positives = 126/205 (61%), Gaps = 28/205 (13%)
Frame = +2
Query: 654 HHLSFPPFSSHLSLLSIFSCPMLTCIPTFPNLDKTLHLVSTSVETLEATLNMVGSELAIE 713
HHL P F LS+L+I C MLT +PTFPN+ K L L +S E LEATLN+ S+ +I
Sbjct: 5 HHLLLPHFPC-LSILTISGCYMLTYMPTFPNI-KILVLRRSSAEILEATLNIAESQYSIG 178
Query: 714 FPPLSKLKYLRLGG-------------------EDLDLKILP---------FFKEDHNFL 745
FPPLS LK L++ EDLD L +FK D N L
Sbjct: 179 FPPLSMLKSLKINETIMGMEKAPKDWFKNLTSLEDLDFDNLSSQGFQVIEMWFKNDPNCL 358
Query: 746 SSIQNFEFCNCSDLKVLPDWICNLSSLQHISIQRCRNLASLPEGMPRLSKLHTLEIFGCP 805
S+Q F CSDLK LPDWICN+SSLQH+++ CR LA LP+ MPRL+ L TLEI CP
Sbjct: 359 RSLQKITFTWCSDLKELPDWICNISSLQHLTVYHCRYLALLPKEMPRLTNLKTLEIIECP 538
Query: 806 LLVEECVTQTSATWSKISHIPNIIL 830
LL+EEC+ +TSAT KI+HIPNI+L
Sbjct: 539 LLIEECLRETSATGPKIAHIPNIVL 613
>BG451530 homologue to GP|23505092|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (4%)
Length = 670
Score = 173 bits (439), Expect = 2e-43
Identities = 93/184 (50%), Positives = 111/184 (59%), Gaps = 28/184 (15%)
Frame = +1
Query: 675 MLTCIPTFPNLDKTLHLVSTSVETLEATLNMVGSELAIEFPPLSKLKYLRLGGEDLDLKI 734
MLT PTFPNL+ L L +SVETL ATLN + E + PPLS LK L + G L++K
Sbjct: 4 MLTSTPTFPNLENGLELFDSSVETLVATLN-IALECLNDIPPLSMLKSLHIDGVSLNVKR 180
Query: 735 LP----------------------------FFKEDHNFLSSIQNFEFCNCSDLKVLPDWI 766
+P +FK+D L S+Q F NC DL+ LPDWI
Sbjct: 181 IPENWMQNLTSLQLLQINWFSRQAFQEIETWFKDDLKCLPSLQTIAFHNCEDLEALPDWI 360
Query: 767 CNLSSLQHISIQRCRNLASLPEGMPRLSKLHTLEIFGCPLLVEECVTQTSATWSKISHIP 826
CNLSSLQH+ + C NLASLPEGMPRL+ L T+EI GCP+LVEEC TQT TW KI H+P
Sbjct: 361 CNLSSLQHLRVYDCINLASLPEGMPRLTNLQTIEIIGCPILVEECQTQTGETWPKIGHVP 540
Query: 827 NIIL 830
IIL
Sbjct: 541 KIIL 552
>TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type putative
resistance protein {Glycine max}, partial (61%)
Length = 676
Score = 172 bits (435), Expect = 6e-43
Identities = 92/228 (40%), Positives = 135/228 (58%)
Frame = +2
Query: 73 VSLVAIVGIGGLGKTALAQLVYNDAQVTKSFEKRMWVCVSDNFDVKTILKKMLESLTNKK 132
VS ++IVG+GG+GKT LAQLVYND ++ ++FE + WV VS+ FDV + +L +
Sbjct: 5 VSTISIVGLGGMGKTTLAQLVYNDRRIQETFELKAWVYVSEYFDVIGLTNAILRKFGTAE 184
Query: 133 IDDKLSLENLQSMLRDTLTAMRYLLVLDDIWNDSFEKWAQLKTYLMCGAQGSKVVVTTRS 192
+ L L LQ L+ LT YLLV+DD+W + E W +L G+ GSK+++TTR
Sbjct: 185 NSEDLDL--LQWQLQKKLTGKNYLLVMDDVWKLNEESWEKLLLPFNYGSSGSKIILTTRD 358
Query: 193 KVVAQTMGVSVPYTLNGLTPEKSWSLLKNIVTYGDETKGVLNQTLETIGKKIAVKCSGVP 252
K VA + + L L + WSL K + +G LE+IGK I KC G+P
Sbjct: 359 KKVALIVKSTELVDLEQLKNKDCWSLFKRLAFHGSNVSEY--PKLESIGKNIVDKCGGLP 532
Query: 253 LAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMPVLKLSYHNL 300
L +T+G LL+ K ++EW +L+ D W+L +++ +I L+LSYHNL
Sbjct: 533 LTGKTMGNLLRKKFTQSEWEKILEADMWRLTDDDSNINSALRLSYHNL 676
>BG587633 similar to GP|20385442|gb| resistance gene analog {Vitis vinifera},
partial (22%)
Length = 677
Score = 157 bits (398), Expect = 1e-38
Identities = 76/169 (44%), Positives = 110/169 (64%)
Frame = +3
Query: 232 VLNQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMP 291
V N TLE IG+K A KC G+P+A +TLGGLL+ K + TEW +L D W L ++I+P
Sbjct: 12 VPNTTLEAIGRKTARKCGGLPIAAKTLGGLLRSKVEITEWTSILNSDIWNL--SNDNILP 185
Query: 292 VLKLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQF 351
L LSY L L++CFAYC+I+PKD+ + + +L+ LWMA+G+L+CS K ME++G+
Sbjct: 186 ALHLSYQYLPCHLKRCFAYCSIFPKDYPLDRKQLVLLWMAEGFLDCSHGGKAMEELGDDC 365
Query: 352 VNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLDSET 400
L +S +Q + D+ G+ F MHDL++DLA ++G C LD T
Sbjct: 366 FAELLSRSLIQQLSNDARGE--KFVMHDLVNDLATVISGQSCFNLDVVT 506
>AW694719 similar to GP|11994217|db disease resistance comples protein
{Arabidopsis thaliana}, partial (6%)
Length = 511
Score = 150 bits (380), Expect = 1e-36
Identities = 75/167 (44%), Positives = 106/167 (62%), Gaps = 3/167 (1%)
Frame = +1
Query: 234 NQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGV---LQDDFWKLCEEEESIM 290
N LE IG+KIA +C G+P+A +TLGGLL+ K D TEW + L W L ++I+
Sbjct: 4 NTALEEIGRKIARRCGGLPIAAKTLGGLLRSKVDITEWTSIFSILNSSIWNL--RNDNIL 177
Query: 291 PVLKLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQ 350
P L LSY L L++CFAYC+I+PKD + K +L+ LWMA+G+L+CS K +E++G+
Sbjct: 178 PALHLSYQYLPSHLKRCFAYCSIFPKDCPLDKKQLVLLWMAEGFLDCSQGGKKLEELGDD 357
Query: 351 FVNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLD 397
L +S +Q + D G+ F MHDLI+DLA V+G CC L+
Sbjct: 358 CFAELLSRSLIQQLSDDDRGE--KFVMHDLINDLATFVSGKSCCRLE 492
>TC83046 weakly similar to GP|10280097|emb|CAC09980. nucleotide sequence of
the I-2 resistance gene {Fusarium oxysporum}, partial
(8%)
Length = 884
Score = 148 bits (374), Expect = 7e-36
Identities = 73/161 (45%), Positives = 103/161 (63%)
Frame = +1
Query: 237 LETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMPVLKLS 296
LE IG+KIA KC G+P+A +TLGG+L+ K D EW +L D W L ++I+P L+LS
Sbjct: 16 LEEIGRKIAKKCGGLPIAAKTLGGILRSKVDAKEWSTILNSDIWNL--PNDNILPALRLS 189
Query: 297 YHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVNIFL 356
Y L L++CFAYC+I+PKD+ + K ELI LWMA+G+LE S K E++G+ + L
Sbjct: 190 YQYLPSHLKRCFAYCSIFPKDFSLDKKELILLWMAEGFLEHSQCNKTAEEVGHDYFIELL 369
Query: 357 MKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLD 397
+S +Q D F MHDL++DLA+ V+G C L+
Sbjct: 370 SRSLIQQSNDDG---KEKFVMHDLVNDLALVVSGTSCFRLE 483
>BE204114 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon
pimpinellifolium}, partial (3%)
Length = 583
Score = 139 bits (350), Expect = 4e-33
Identities = 77/180 (42%), Positives = 108/180 (59%), Gaps = 1/180 (0%)
Frame = +3
Query: 234 NQTLETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMPVL 293
++ L IGK+I KC VPLA LG LL+ K +E EW+ V + W L E E+ + L
Sbjct: 36 DEKLVVIGKEILKKCV*VPLAAIALGSLLRFKREEKEWLYVKESKLWSL-EGEDYVKSAL 212
Query: 294 KLSYHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVN 353
KLSY NL +LRQCF++CA++PKD + KH +I LW+A G++ S + E +GN+ N
Sbjct: 213 KLSYLNLPVKLRQCFSFCALFPKDEIMSKHFMIELWIANGFIS-SNQMLDAEGVGNEVWN 389
Query: 354 IFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGND-CCYLDSETKNLVESPMHIMM 412
+SF QD ETD G I SFKMHDL+H+LA V C +++ + ES H+ +
Sbjct: 390 ELYWRSFFQDTETDEFGQITSFKMHDLVHELAESVTREVWCITYNNDLPTVSESIRHLFI 569
>BM779031 weakly similar to GP|6606266|gb| Vrga1 {Aegilops ventricosa},
partial (4%)
Length = 532
Score = 137 bits (346), Expect = 1e-32
Identities = 75/166 (45%), Positives = 101/166 (60%)
Frame = +2
Query: 344 MEDIGNQFVNIFLMKSFLQDVETDSCGDIHSFKMHDLIHDLAMEVAGNDCCYLDSETKNL 403
+ED+GNQ+V I L++SF QD DI SFKMHDL+HDLA VAGNDCC L +E K
Sbjct: 26 IEDVGNQYVRILLLRSFFQDASLSKHSDIISFKMHDLMHDLAKSVAGNDCC-LHAEGKRF 202
Query: 404 VESPMHIMMKMDDIGLLESVDASRLRTLILMPNLKTFRNEEDMSIISKFKYLRVLKLSHC 463
+ P+H+ L+ +DAS+ RT++ ++ +SI KYLR LSH
Sbjct: 203 IGRPIHVSFLSSTTCSLDLLDASKPRTVLWAKTKAGSVSDAKLSITKNLKYLRAFDLSHS 382
Query: 464 SLCKLCDSIVKLKHLRYLDLWYCRGVGSVFKSITNMVCLQTLKLVG 509
+ +L SI K +HLRYLDL C+ + S+ KSI N+ LQ+LKL G
Sbjct: 383 LITELPQSIDKCRHLRYLDLSSCKELISLPKSIGNLASLQSLKLSG 520
Score = 35.4 bits (80), Expect = 0.091
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Frame = +2
Query: 745 LSSIQNFEFCNCSDLK-----VLPDWICNLSSLQHISIQRCRNLASLPEGMPRLSKLHTL 799
LS +N ++ DL LP I L+++ + C+ L SLP+ + L+ L +L
Sbjct: 329 LSITKNLKYLRAFDLSHSLITELPQSIDKCRHLRYLDLSSCKELISLPKSIGNLASLQSL 508
Query: 800 EIFGC 804
++ GC
Sbjct: 509 KLSGC 523
Score = 32.7 bits (73), Expect = 0.59
Identities = 20/64 (31%), Positives = 32/64 (49%)
Frame = +2
Query: 720 LKYLRLGGEDLDLKILPFFKEDHNFLSSIQNFEFCNCSDLKVLPDWICNLSSLQHISIQR 779
LKYLR DL ++ + + ++ + +C +L LP I NL+SLQ + +
Sbjct: 347 LKYLR--AFDLSHSLITELPQSIDKCRHLRYLDLSSCKELISLPKSIGNLASLQSLKLSG 520
Query: 780 CRNL 783
CR L
Sbjct: 521 CRQL 532
>BG453139 similar to GP|5817349|gb| putative NBS-LRR type disease resistance
protein {Pisum sativum}, partial (65%)
Length = 638
Score = 137 bits (346), Expect = 1e-32
Identities = 78/193 (40%), Positives = 120/193 (61%), Gaps = 2/193 (1%)
Frame = -2
Query: 1 MAHEIEKLQTKFNDVVKDMPGLNLNSNVVVVEQSDIVRRETSSFVLESEIIGREDDKKKI 60
M IE+LQ KD+ GL S V +R +SS V +S ++GR+ DK+K+
Sbjct: 562 MKSMIERLQLFAQQ--KDILGLQTVSGRV------FLRTPSSSLVDQSFMVGRKSDKEKL 407
Query: 61 ISLLRQ--SHENQNVSLVAIVGIGGLGKTALAQLVYNDAQVTKSFEKRMWVCVSDNFDVK 118
++L S N N+++V+I G+GG+GKTALAQL+YND V + F+ + WVC+S+ FDV
Sbjct: 406 TNMLLSDVSTVNDNINVVSIFGMGGVGKTALAQLLYNDKDVQERFDVKAWVCLSEEFDVL 227
Query: 119 TILKKMLESLTNKKIDDKLSLENLQSMLRDTLTAMRYLLVLDDIWNDSFEKWAQLKTYLM 178
+ + +LES+T+ + + +L+ L+ L+ L R+L VLDD+WNDS+ W L T +
Sbjct: 226 RVTRTLLESITS-NVQESDNLDFLRVKLKQNLKGKRFLFVLDDMWNDSYNDWHHLVTPFI 50
Query: 179 CGAQGSKVVVTTR 191
G GS+V++TTR
Sbjct: 49 DGESGSRVIITTR 11
>BG648625 weakly similar to GP|20385440|gb| resistance gene analog {Vitis
vinifera}, partial (21%)
Length = 521
Score = 135 bits (341), Expect = 5e-32
Identities = 67/147 (45%), Positives = 95/147 (64%)
Frame = +3
Query: 237 LETIGKKIAVKCSGVPLAIRTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMPVLKLS 296
L IG+K+ KC G PLA + LG LL+ KSDE +W V++ +FW L ++ +M L+LS
Sbjct: 90 LVEIGQKLVRKCVGSPLAAKVLGSLLRFKSDEHQWTSVVESEFWNLADDNH-VMSALRLS 266
Query: 297 YHNLSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVNIFL 356
Y NL LR CF +CA++PKD+++ K I LWMA G L S ME +GN+ N
Sbjct: 267 YFNLKLSLRPCFTFCAVFPKDFEMEKEFFIQLWMANG-LVTSRGNLQMEHVGNEVWNELY 443
Query: 357 MKSFLQDVETDSCGDIHSFKMHDLIHD 383
+SF Q++++D G+I +FKMHDL+HD
Sbjct: 444 QRSFFQEIKSDLVGNI-TFKMHDLVHD 521
>TC93188
Length = 669
Score = 129 bits (325), Expect = 4e-30
Identities = 67/137 (48%), Positives = 80/137 (57%), Gaps = 28/137 (20%)
Frame = +1
Query: 713 EFPPLSKLKYLRLGGEDLDLKILP----------------------------FFKEDHNF 744
+FPPLS LK L + G LD+K +P +FK+D +
Sbjct: 19 DFPPLSMLKSLHIDGVRLDVKSIPNVWMKNLTSLQLLQINWFSRQAFQQIETWFKDDLKY 198
Query: 745 LSSIQNFEFCNCSDLKVLPDWICNLSSLQHISIQRCRNLASLPEGMPRLSKLHTLEIFGC 804
L S+Q F NC DL+ LPDWICNLSSLQH+ + C NLASLPEGM L+ L TLEI GC
Sbjct: 199 LPSLQTIAFHNCEDLEALPDWICNLSSLQHLRVYDCINLASLPEGMLNLTNLQTLEIIGC 378
Query: 805 PLLVEECVTQTSATWSK 821
P+LVEEC TQT TW K
Sbjct: 379 PILVEECQTQTGETWDK 429
>BE943049
Length = 675
Score = 129 bits (323), Expect = 6e-30
Identities = 68/144 (47%), Positives = 87/144 (60%), Gaps = 28/144 (19%)
Frame = +1
Query: 715 PPLSKLKYLRLGGEDLDL----------------------------KILPFFKEDHNFLS 746
PPLS LK L +GG L + +I +F + N L
Sbjct: 1 PPLSMLKSLCIGGHKLAVYNISENWMQNLPSLQHLQIELFSSQQVHEIAIWFNNNFNCLP 180
Query: 747 SIQNFEFCNCSDLKVLPDWICNLSSLQHISIQRCRNLASLPEGMPRLSKLHTLEIFGCPL 806
S+Q C DLK LPDW+C++SSLQH++I+ +LAS+PEGMPRL+KL TLEI GCPL
Sbjct: 181 SLQKITLQYCDDLKALPDWMCSISSLQHVTIRYSPHLASVPEGMPRLAKLKTLEIIGCPL 360
Query: 807 LVEECVTQTSATWSKISHIPNIIL 830
LV+EC QT+ATW K++HIPNIIL
Sbjct: 361 LVKECEAQTNATWPKVAHIPNIIL 432
>BG451218 similar to GP|11994217|db disease resistance comples protein
{Arabidopsis thaliana}, partial (5%)
Length = 666
Score = 113 bits (282), Expect(2) = 2e-28
Identities = 60/155 (38%), Positives = 89/155 (56%)
Frame = +1
Query: 256 RTLGGLLQGKSDETEWVGVLQDDFWKLCEEEESIMPVLKLSYHNLSPQLRQCFAYCAIYP 315
+T+GGLL K D EW +L + W L + I+P L LSY L L+ CFAYC+I+P
Sbjct: 88 KTIGGLLGSKVDIIEWTTILNSNVWNL--PNDKILPTLHLSYQCLPSHLKICFAYCSIFP 261
Query: 316 KDWKIHKHELIHLWMAQGYLECSAKKKLMEDIGNQFVNIFLMKSFLQDVETDSCGDIHSF 375
K + +L+ LWMA+G+L+ S +K ME++G+ L +S +Q + G+ F
Sbjct: 262 KGHTHDRKKLVLLWMAEGFLDYSHGEKTMEELGDDCFAELLSRSLIQQSNDNGRGE--KF 435
Query: 376 KMHDLIHDLAMEVAGNDCCYLDSETKNLVESPMHI 410
MHDL++DLA V+G CC E N+ E+ H+
Sbjct: 436 FMHDLVNDLATVVSGKSCCRF--ECGNISENVRHV 534
Score = 32.0 bits (71), Expect(2) = 2e-28
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +3
Query: 234 NQTLETIGKKIAVKCSGVPLA 254
N T E IG+KIA KC G+P+A
Sbjct: 21 NSTFEEIGRKIARKCGGLPIA 83
>BF639547 similar to GP|15553678|gb I2C-5 {Lycopersicon pimpinellifolium},
partial (4%)
Length = 660
Score = 120 bits (301), Expect = 2e-27
Identities = 82/193 (42%), Positives = 113/193 (58%), Gaps = 9/193 (4%)
Frame = +1
Query: 300 LSPQLRQCFAYCAIYPKDWKIHKHELIHLWMAQGYLECSAKKKL-MEDIGNQFVNIFLMK 358
L+P L+QCF++CAI+PKD +I K ELI LWMA G++ S+K L +ED+GN K
Sbjct: 73 LTPTLKQCFSFCAIFPKDGEILKEELIQLWMANGFI--SSKGNLDVEDVGNMVWKELYQK 246
Query: 359 SFLQDVETDS-CGDIHSFKMHDLIHDLAMEVAGNDCCYLD-SETKNLVESPMHIMMKMD- 415
SF QD++ D GDI FKMHDL+HDLA V G +C YL+ + +L +S HI D
Sbjct: 247 SFFQDIKMDEYSGDIF-FKMHDLVHDLAQSVMGQECVYLENANMTSLTKSTHHISFNSDN 423
Query: 416 ----DIGLLESVDASRLRTLIL-MPNLKTFRNEEDMSIISKFKYLRVLKLSHCSLCKLCD 470
D G + V++ LRTL+ + N F + D ++ + LRVL +SH
Sbjct: 424 LLSFDEGAFKKVES--LRTLLFNLKNPNFFAKKYDHFPLN--RSLRVLCISHVL------ 573
Query: 471 SIVKLKHLRYLDL 483
S+ L HLRYL+L
Sbjct: 574 SLESLIHLRYLEL 612
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,222,818
Number of Sequences: 36976
Number of extensions: 476788
Number of successful extensions: 3165
Number of sequences better than 10.0: 240
Number of HSP's better than 10.0 without gapping: 2917
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3046
length of query: 831
length of database: 9,014,727
effective HSP length: 104
effective length of query: 727
effective length of database: 5,169,223
effective search space: 3758025121
effective search space used: 3758025121
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126793.6


