

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.9 + phase: 0 /pseudo
(1040 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
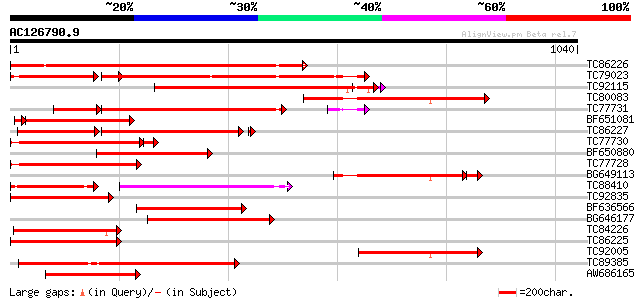
Score E
Sequences producing significant alignments: (bits) Value
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 772 0.0
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 630 0.0
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 525 e-149
TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 458 e-129
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 455 e-128
BF651081 weakly similar to GP|18033111|g functional candidate re... 369 e-110
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 390 e-109
TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 334 4e-99
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 358 5e-99
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 333 3e-91
BG649113 similar to GP|18033111|gb functional candidate resistan... 314 3e-91
TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 189 2e-85
TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 313 3e-85
BF636566 weakly similar to GP|9965103|gb| resistance protein LM6... 309 3e-84
BG646177 weakly similar to GP|18033111|g functional candidate re... 305 5e-83
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 302 5e-82
TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance p... 293 2e-79
TC92005 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 290 3e-78
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 276 4e-74
AW686165 weakly similar to GP|18033111|gb functional candidate r... 274 1e-73
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 772 bits (1994), Expect = 0.0
Identities = 401/548 (73%), Positives = 450/548 (81%), Gaps = 2/548 (0%)
Frame = +3
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNG--LQRGDE 58
MQ P S+S SY + Y VFL +G+DTRYGFTGNL KAL DKGI TF D + LQR D+
Sbjct: 45 MQLPFYSSSFSYAFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDK 224
Query: 59 ITPSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVR 118
+TP + IEESRI IP+FS NYASSS CLD LVHIIHCYKTKG LVLPVFFGVEPT VR
Sbjct: 225 VTPII---IEESRILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVR 395
Query: 119 HRKGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVK 178
H G YG+ALAEHE RFQND KNMERLQ WK ALS AANL YHD GYEY+LIGKIVK
Sbjct: 396 HHTGRYGKALAEHENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVK 575
Query: 179 YISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIY 238
YISNKISRQ LHVATYPVGLQSRVQQVKSLLDEG DDGVHMVGIYGIGG GKSTLAR IY
Sbjct: 576 YISNKISRQSLHVATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIY 755
Query: 239 NFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRK 298
NFVADQFEG CFL VRENSA N+LK QE LL KT L+IKL VSEGI +IKERLCRK
Sbjct: 756 NFVADQFEGLCFLEQVRENSASNSLKRFQEMLLSKTLQLKIKLADVSEGISIIKERLCRK 935
Query: 299 KILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEA 358
KILLILDDVDN+KQL+ALAGG+DWFG GSRVIITTR+K LL+ H IE T+AV+GLN TEA
Sbjct: 936 KILLILDDVDNMKQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEA 1115
Query: 359 LELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDR 418
LELLRWMAFK+DKVPS YE ILNR VAYA GLP+V+E+VGSNLFGK+IE+ K+TLD Y++
Sbjct: 1116LELLRWMAFKNDKVPSSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEK 1295
Query: 419 IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLA 478
IPNKEIQ+ILKVSYD+LEEEEQSVFLDIACCFKG +W++ ++IL AHY HCI HH+ VL
Sbjct: 1296IPNKEIQRILKVSYDSLEEEEQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLV 1475
Query: 479 GKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKEN 538
K L+ Y + V LH+LI++MGKE+VR ESP EPG+RSRLW ++DI VL+EN
Sbjct: 1476EKCLIDHFEY-----DSHVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEEN 1640
Query: 539 TGTSKIEM 546
T SKI+M
Sbjct: 1641T-VSKIDM 1661
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 630 bits (1625), Expect = 0.0
Identities = 334/492 (67%), Positives = 380/492 (76%)
Frame = +3
Query: 168 YEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGG 227
YEY+ IGKIV+ ISN+ISR+PL VA YPVGLQSRVQ VK LDE SDD VHMVG+YG GG
Sbjct: 690 YEYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGG 869
Query: 228 LGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEG 287
+GKSTLA+ IYNF+ADQFE CFL +VR NS +NLK+LQEKLLLKT L+IKL VS+G
Sbjct: 870 IGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQG 1049
Query: 288 IPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIEST 347
IP+IK+RLCRKKILLILDDVD L QL ALAGGLDWFG GSRVIITTRNK LL HGIEST
Sbjct: 1050 IPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIEST 1229
Query: 348 HAVEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIE 407
HAVEGLN TEALELLRWMAFK + VPS +EDILNRA+ YA GLPL + ++GSNL G+S++
Sbjct: 1230 HAVEGLNATEALELLRWMAFK-ENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQ 1406
Query: 408 DWKHTLDGYDRIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYD 467
D TLDGY+ IPNKEIQ+ILKVSYD+LE+EEQSVFLDIACCFKG +W E ++IL AHY
Sbjct: 1407 DSMSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYG 1586
Query: 468 HCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWR 527
HCI HH+ VLA KSL+ Y + V LHDLI+DMGKEVVRQESP EPGERSRLW
Sbjct: 1587 HCIVHHVAVLAEKSLMDHLKY-----DSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWF 1751
Query: 528 QEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKY 587
+ DI+HVLK+NTGT KI+MI M SMES ID G AF+KMT LKT I ENG S L+Y
Sbjct: 1752 ERDIVHVLKKNTGTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEY 1931
Query: 588 LPSSLRVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDY 647
LPSSLRV+ KGC+ K SSS NK KF++MKVL L+
Sbjct: 1932 LPSSLRVM--KGCIPKSPSSSSSNK-----------------------KFEDMKVLILNN 2036
Query: 648 CEYLTHIPDVSG 659
CEYLTHIPDVSG
Sbjct: 2037 CEYLTHIPDVSG 2072
Score = 245 bits (626), Expect(2) = 5e-72
Identities = 126/161 (78%), Positives = 133/161 (82%)
Frame = +1
Query: 2 QSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITP 61
QSPSS T YQVFLSFRG+DTR+GFTGNLYKALTDKGI TFID N LQRGDEITP
Sbjct: 73 QSPSSFT-------YQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITP 231
Query: 62 SLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRK 121
SL AIE+SRIFIPVFS NYASSSFCLDELVHI HCY TKG LVLPVF GV+PT VRH
Sbjct: 232 SLKNAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHT 411
Query: 122 GSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH 162
G YGEALA H+K+FQND N ERLQ WK+ALSQAANLSG H
Sbjct: 412 GRYGEALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQH 534
Score = 45.4 bits (106), Expect(2) = 5e-72
Identities = 23/41 (56%), Positives = 31/41 (75%)
Frame = +2
Query: 169 EYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLL 209
EY+ IGKIV+ ISN+I+R+P VA Y V LQ +VQ +K+ L
Sbjct: 551 EYEFIGKIVEDISNRINRKPQDVAKYHVELQLQVQHIKTHL 673
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 525 bits (1352), Expect = e-149
Identities = 293/439 (66%), Positives = 334/439 (75%), Gaps = 28/439 (6%)
Frame = +2
Query: 266 LQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGC 325
LQE+LLLK L+IKL VSEGIP IKERL KK LLILDDVD+++QLHALAGG DWFG
Sbjct: 2 LQEELLLKALQLKIKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFGR 181
Query: 326 GSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVA 385
GSRVIITTR+K LL SHGIESTH VE L TEALELLRWMAFK++KVPS YED+LNRAV+
Sbjct: 182 GSRVIITTRDKHLLRSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSIYEDVLNRAVS 361
Query: 386 YAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYDALEEEEQSVFLD 445
YA GLPLVLE+VGSNLFGK+IE+WK TLDGY++IPNK+I +ILKVSYDALEEE+QSVFLD
Sbjct: 362 YASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEEQQSVFLD 541
Query: 446 IACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKD 505
IACCFKG W+EFE IL AHY H ITHHL VLA KSLVKI T+ GSI ++ LHDLIK+
Sbjct: 542 IACCFKGCGWEEFEYILRAHYGHRITHHLVVLAEKSLVKI-THPHYGSIYELTLHDLIKE 718
Query: 506 MGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAF 565
MGKEVVRQESPKEPGERSRLW ++DI++VLKENTGTSKIEMIYMN S E VIDKKGKAF
Sbjct: 719 MGKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNFPSEEFVIDKKGKAF 898
Query: 566 KKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSILNKASEITS------- 618
KKMT+LKTLIIEN FS GLKYLPSSLRVLK +GCLS+ L S L+KA EITS
Sbjct: 899 KKMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRGCLSESLISCSLSKAIEITSFSYCIYL 1078
Query: 619 ---Q*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPD------------------V 657
FCF QQ+++ + N ++ L +P+
Sbjct: 1079LIYNDFCFRQQRRNRYDSI--VFNF*LIYLFLPXVYAEVPEHENLDIGQV*IF*PIYLMX 1252
Query: 658 SGLSNLEKLSFTCCDNLIT 676
SGL NL+K SF +NLIT
Sbjct: 1253SGLQNLKKFSFEYXENLIT 1309
Score = 47.0 bits (110), Expect = 4e-05
Identities = 28/60 (46%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Frame = +3
Query: 630 FLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEKLSFTCCDNL-ITIHNSIGHLNKLE 688
FL MQKFQNMK+LTLD CEY + + K S + + IHNSIGHL L+
Sbjct: 1167 FLXFMQKFQNMKILTLDRCEYFNPYT*CXQVFKI*KSSPLNIXRI*LPIHNSIGHLINLK 1346
>TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (4%)
Length = 1269
Score = 458 bits (1179), Expect = e-129
Identities = 250/352 (71%), Positives = 273/352 (77%), Gaps = 11/352 (3%)
Frame = +3
Query: 540 GTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKG 599
GTSKIEMIYMNLHSMESVID+KGKAFKKMTKLKTLIIENG S GLKYLPSSL VLKWKG
Sbjct: 18 GTSKIEMIYMNLHSMESVIDEKGKAFKKMTKLKTLIIENGHCSKGLKYLPSSLSVLKWKG 197
Query: 600 CLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSG 659
CLSKCLSS ILNK KFQ+MKVL LD C+YLTHIPDVSG
Sbjct: 198 CLSKCLSSRILNK-----------------------KFQDMKVLILDRCKYLTHIPDVSG 308
Query: 660 LSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECEC 719
LSNLEKLSF C NLITIHNSIGHLNKLE LSAYGC K+EHF PLGLASLK+L L C
Sbjct: 309 LSNLEKLSFERCYNLITIHNSIGHLNKLERLSAYGCNKVEHFPPLGLASLKELNLSSCRS 488
Query: 720 LDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGM-KFPK-------IV 771
L +FPELLCKM +I I + NTSIGELPFSFQNLSELH+L+VT GM +FPK IV
Sbjct: 489 LKSFPELLCKMTNIDNIWLCNTSIGELPFSFQNLSELHKLSVTYGMLRFPKQNDKMYSIV 668
Query: 772 FSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCC 831
FSN+T+L+L NLSDECL VLKWCVNMT L+L +SNFKILPECL ECHHLVEINV C
Sbjct: 669 FSNVTQLTLYHCNLSDECLRRVLKWCVNMTSLNLLYSNFKILPECLSECHHLVEINVGYC 848
Query: 832 ESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQP--SITCIFI-LPKGNE 880
ESLEEIRGIPPNLKE+ A+ C+SLSSSS+R+LMSQ C +I P G +
Sbjct: 849 ESLEEIRGIPPNLKEINAQGCQSLSSSSKRILMSQKLHQAGCTYIKFPNGTD 1004
Score = 32.7 bits (73), Expect = 0.74
Identities = 24/54 (44%), Positives = 32/54 (58%)
Frame = +2
Query: 867 PSITCIFILPKGNEYATSVNVFVNGYEIEIGCYWSLFFFTDHTTLFHTSKLNEL 920
PSIT I I P+ + ++V ++VNGY IEI Y + + TTL H KL EL
Sbjct: 1076 PSITGIIIFPEC-AWGSNVGLYVNGY*IEID-YCYQYVSSKDTTLIH-MKLYEL 1228
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 455 bits (1171), Expect = e-128
Identities = 232/340 (68%), Positives = 271/340 (79%)
Frame = +1
Query: 168 YEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGG 227
YEYK I KIV+ ISN I+ L+VA YPVGLQSR++QVK LLD GS+D V MVG++G GG
Sbjct: 499 YEYKFIEKIVEDISNNINHVFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGG 678
Query: 228 LGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEG 287
+GKSTLA+ +YNFVADQFEG CFLH+VRENS NLK+LQ+KLL K + KL+ VSEG
Sbjct: 679 MGKSTLAKAVYNFVADQFEGVCFLHNVRENSTHKNLKHLQKKLLSKIVKFDGKLEDVSEG 858
Query: 288 IPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIEST 347
IP+IKERL RKKILLILDDVD L+QL ALAGGLDWFG GSRVIITTR+K LL+ HGI ST
Sbjct: 859 IPIIKERLSRKKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLACHGITST 1038
Query: 348 HAVEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIE 407
HAVE LNETEALELLR MAFK+DKVPS YE+ILNR V YA GLPL + +G NLFG+ +E
Sbjct: 1039HAVEELNETEALELLRRMAFKNDKVPSTYEEILNRVVTYASGLPLAIVTIGDNLFGRKVE 1218
Query: 408 DWKHTLDGYDRIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYD 467
DWK LD Y+ IPNK+IQ+IL+VSYDALE +E+SVFLDIACCFKG +W + + IL AHY
Sbjct: 1219DWKRILDEYENIPNKDIQRILQVSYDALEPKEKSVFLDIACCFKGCKWTKVKKILHAHYG 1398
Query: 468 HCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMG 507
HCI HH+GVLA KSL+ Y + LHDLI+DMG
Sbjct: 1399HCIEHHVGVLAEKSLIGHWEY-----DTQMTLHDLIEDMG 1503
Score = 144 bits (364), Expect = 1e-34
Identities = 68/88 (77%), Positives = 76/88 (86%)
Frame = +2
Query: 81 YASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRFQNDPK 140
YASSSFCLDELVHIIHCYKTK LVLPVF+ VEPT +RH+ GSYGE L +HE+RFQN+ K
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 141 NMERLQGWKKALSQAANLSGYHDSPPGY 168
NMERL+ WK AL+QAANLSGYH SP GY
Sbjct: 182 NMERLRQWKIALTQAANLSGYHYSPHGY 265
Score = 43.5 bits (101), Expect = 4e-04
Identities = 33/76 (43%), Positives = 36/76 (46%)
Frame = +3
Query: 584 GLKYLPSSLRVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVL 643
G YLPSSLR L+W K LS IL+K +F MKVL
Sbjct: 1494 GHGYLPSSLRYLEWIDYDFKSLSC-ILSK-----------------------EFNYMKVL 1601
Query: 644 TLDYCEYLTHIPDVSG 659
LDY LTHIPDVSG
Sbjct: 1602 KLDYSSDLTHIPDVSG 1649
>BF651081 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (16%)
Length = 664
Score = 369 bits (948), Expect(2) = e-110
Identities = 181/199 (90%), Positives = 189/199 (94%)
Frame = +1
Query: 30 GFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIPVFSINYASSSFCLD 89
GFTGNLYKAL DKGI+TFID + LQRGDEITPSLLKAIEESRIFI VFSINYASSSFCLD
Sbjct: 67 GFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYASSSFCLD 246
Query: 90 ELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRFQNDPKNMERLQGWK 149
ELVHIIHCYKTKGRLVLPVFF VEPT+VRH+KGSYGEALAEHEKRFQNDPK+MERLQGWK
Sbjct: 247 ELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSMERLQGWK 426
Query: 150 KALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLL 209
+ALSQAANLSGYHDSPPGYEYKLIGKIVKYI NKIS+QPLHVATYPVGLQS QQ+KSLL
Sbjct: 427 EALSQAANLSGYHDSPPGYEYKLIGKIVKYIXNKISQQPLHVATYPVGLQSXXQQMKSLL 606
Query: 210 DEGSDDGVHMVGIYGIGGL 228
DE SD GVHMVGIYGIGGL
Sbjct: 607 DEXSDHGVHMVGIYGIGGL 663
Score = 47.8 bits (112), Expect(2) = e-110
Identities = 21/23 (91%), Positives = 22/23 (95%)
Frame = +3
Query: 9 SISYEYKYQVFLSFRGSDTRYGF 31
SISYEYKYQVFL+FRGSDTRY F
Sbjct: 3 SISYEYKYQVFLNFRGSDTRYXF 71
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 390 bits (1001), Expect(2) = e-109
Identities = 197/262 (75%), Positives = 226/262 (86%)
Frame = +1
Query: 168 YEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGG 227
YEY+LI IV++IS++I+R LHVA YPVGLQSRVQQVK LLDE SD+GV+MVG+YG
Sbjct: 682 YEYELIENIVEHISDRINRVFLHVAKYPVGLQSRVQQVKLLLDEESDEGVNMVGLYGTRA 861
Query: 228 LGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEG 287
LGKSTLA+ IYNF+ADQFEG CFLH+VRENSA+ NLK+LQ++LL KT L IKL VSEG
Sbjct: 862 LGKSTLAKAIYNFIADQFEGVCFLHNVRENSARKNLKHLQKELLSKTVQLNIKLRDVSEG 1041
Query: 288 IPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIEST 347
IP+IKERLCRKKILLILDDVD L QL ALAGGLDWFG GSRVIITTR+K LL+ HGIE T
Sbjct: 1042IPIIKERLCRKKILLILDDVDQLDQLEALAGGLDWFGPGSRVIITTRDKHLLTCHGIERT 1221
Query: 348 HAVEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIE 407
+AV GL EALELLRW AFK++KVP YED+LNRAV+Y G+PLVLE+VGSNLFGK+IE
Sbjct: 1222YAVRGLYGKEALELLRWTAFKNNKVPPSYEDVLNRAVSYGSGIPLVLEIVGSNLFGKNIE 1401
Query: 408 DWKHTLDGYDRIPNKEIQKILK 429
WK+TLDGYDRIPNKEIQKIL+
Sbjct: 1402VWKNTLDGYDRIPNKEIQKILR 1467
Score = 233 bits (594), Expect = 3e-61
Identities = 112/151 (74%), Positives = 127/151 (83%)
Frame = +3
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIF 73
+KY VFLSFR DT YGFTGNLYKAL DKGI TFID N L+RGDE TPSL+KAIEESRI
Sbjct: 12 FKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEESRIL 191
Query: 74 IPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEK 133
IP+FS NYASSSFCLDELVHIIHCYKT+G VLPVF+G +PT VRH+ GSYGE L +HE
Sbjct: 192 IPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTKHED 371
Query: 134 RFQNDPKNMERLQGWKKALSQAANLSGYHDS 164
+FQN+ +NMERL+ WK AL+QAAN SG+H S
Sbjct: 372 KFQNNKENMERLKKWKMALTQAANFSGHHFS 464
Score = 26.6 bits (57), Expect(2) = e-109
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +2
Query: 438 EEQSVFLDIACCF 450
EEQ+VFLDIAC F
Sbjct: 1466 EEQNVFLDIACFF 1504
>TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (80%)
Length = 831
Score = 334 bits (856), Expect(2) = 4e-99
Identities = 170/246 (69%), Positives = 197/246 (79%)
Frame = +1
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
MQSPS +VFL+FRGSDTR FTGNLYKAL DKGI TFID+N LQRGDEIT
Sbjct: 22 MQSPS-----------RVFLNFRGSDTRNNFTGNLYKALVDKGIRTFIDENDLQRGDEIT 168
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHR 120
PSL+KAIEESRI IP+FS NYASSSFCLDELVHIIHCYKTK LVLPVF+GV+PT VR
Sbjct: 169 PSLVKAIEESRICIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYGVDPTDVRRH 348
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYI 180
SYGE L +HE+ FQN+ KNMERL+ WK AL+QAANLSGYH SP YE+K I KIV+YI
Sbjct: 349 TCSYGEYLTKHEEGFQNNEKNMERLRQWKMALTQAANLSGYHYSPHEYEHKFIEKIVQYI 528
Query: 181 SNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNF 240
SN I+ L+VA YPVGLQSR++QVK LLD GS+D V MVG++G GG+GKSTLA+ ++NF
Sbjct: 529 SNNINHDFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVFNF 708
Query: 241 VADQFE 246
+AD +
Sbjct: 709 LADHLK 726
Score = 47.4 bits (111), Expect(2) = 4e-99
Identities = 21/29 (72%), Positives = 25/29 (85%)
Frame = +3
Query: 245 FEGSCFLHDVRENSAQNNLKYLQEKLLLK 273
FEG CFLH+VRENS+ NNLK+LQ+ LL K
Sbjct: 720 FEGVCFLHNVRENSSHNNLKHLQKMLLSK 806
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 358 bits (920), Expect = 5e-99
Identities = 180/212 (84%), Positives = 193/212 (90%)
Frame = +3
Query: 160 GYHDSPPGYEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHM 219
GYHDSPP YEYKLIG+IVKYISNKI+RQPLHVA YPVGL SRVQ+VKSLLDEG DDGVHM
Sbjct: 3 GYHDSPPRYEYKLIGEIVKYISNKINRQPLHVANYPVGLHSRVQEVKSLLDEGPDDGVHM 182
Query: 220 VGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEI 279
VGIYGIGGLGKS LAR IYNFVADQFEG CFLHDVRENSAQNNLK+LQEKLLLKTTGL+I
Sbjct: 183 VGIYGIGGLGKSALARAIYNFVADQFEGLCFLHDVRENSAQNNLKHLQEKLLLKTTGLKI 362
Query: 280 KLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLL 339
KLDHV EGIP+IKERLCR KILLILDDVD+++QLHALAGG DWFG GSRVIITTR+K LL
Sbjct: 363 KLDHVCEGIPIIKERLCRNKILLILDDVDDMEQLHALAGGPDWFGHGSRVIITTRDKHLL 542
Query: 340 SSHGIESTHAVEGLNETEALELLRWMAFKSDK 371
+SH IE T+AVEGL T+ALELLR MAFK K
Sbjct: 543 TSHDIERTYAVEGLYGTKALELLRXMAFKITK 638
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 333 bits (853), Expect = 3e-91
Identities = 170/241 (70%), Positives = 194/241 (79%)
Frame = +1
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
MQSPS +VFLSFRGSDTR FTGNLYKAL DKGI TFID N L+RGDEIT
Sbjct: 76 MQSPS-----------RVFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGDEIT 222
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHR 120
PSL+KAIEESRIFIP+FS NYASSSFCLDELVHIIHCYKTK LV PVF+ VEPT +R++
Sbjct: 223 PSLVKAIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHIRNQ 402
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYI 180
G YGE L +HE+RFQN+ KNMERL+ WK AL QAANLSGYH SP GYEYK I KIV+ I
Sbjct: 403 SGIYGEHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHGYEYKFIEKIVEDI 582
Query: 181 SNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNF 240
SN I+ L+VA YPVGLQSR+++VK LLD GS+D V MVG++G GG+GKSTLA+ +YNF
Sbjct: 583 SNNINHVFLNVAKYPVGLQSRIEEVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVYNF 762
Query: 241 V 241
V
Sbjct: 763 V 765
>BG649113 similar to GP|18033111|gb functional candidate resistance protein
KR1 {Glycine max}, partial (2%)
Length = 782
Score = 314 bits (804), Expect(2) = 3e-91
Identities = 171/253 (67%), Positives = 190/253 (74%), Gaps = 8/253 (3%)
Frame = +1
Query: 595 LKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHI 654
++ +GCLSKCLS+SILNK KFQNMK+LTLD CEYLTHI
Sbjct: 4 IEMEGCLSKCLSTSILNK-----------------------KFQNMKILTLDDCEYLTHI 114
Query: 655 PDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLIL 714
PDVS LSNLEKLSF C NLITIHNSIGHL+KLE LS G RKL+HF PLGLASLK+L L
Sbjct: 115 PDVSSLSNLEKLSFEHCKNLITIHNSIGHLSKLERLSVTGYRKLKHFPPLGLASLKELNL 294
Query: 715 YECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSG-MKFPK---- 769
CL+NFPELLCKMAHIKEIDI SIG+LPFSFQNLSEL E TV+ G ++FP+
Sbjct: 295 MGGSCLENFPELLCKMAHIKEIDIFYISIGKLPFSFQNLSELDEFTVSYGILRFPEHNDK 474
Query: 770 ---IVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSNFKILPECLRECHHLVEI 826
IVFSNMTKLSL LSDECLPI+LKWCVNMT+LDLS+S+FKILPECL E HHLVEI
Sbjct: 475 MYSIVFSNMTKLSLFDCYLSDECLPILLKWCVNMTYLDLSYSDFKILPECLSESHHLVEI 654
Query: 827 NVMCCESLEEIRG 839
V C+SLEEI G
Sbjct: 655 IVRYCKSLEEIEG 693
Score = 41.2 bits (95), Expect(2) = 3e-91
Identities = 21/29 (72%), Positives = 22/29 (75%)
Frame = +3
Query: 838 RGIPPNLKELCARYCKSLSSSSRRMLMSQ 866
RGIPPNL L A CKSLSSS RR +MSQ
Sbjct: 687 RGIPPNLGSLYAYECKSLSSSCRRNVMSQ 773
>TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (8%)
Length = 2252
Score = 189 bits (481), Expect(2) = 2e-85
Identities = 122/324 (37%), Positives = 185/324 (56%), Gaps = 6/324 (1%)
Frame = +1
Query: 201 RVQQVKSLLDEG---SDDGVHMVGIYGIGGLGKSTLARQIYNFVAD-QFEGSCFLHDVRE 256
RV++V L+ DDGV +VGI G+ G+GK+TLAR +Y+F +F+ CF +V E
Sbjct: 604 RVEKVMRYLNSSPRSDDDGVCLVGICGVPGIGKTTLARGVYHFGGGTEFDSCCFFDNVGE 783
Query: 257 NSAQNNLKYLQEKLLLKTTGLE--IKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLH 314
++ L +LQ+ LL G ++V E + IK L +KK+ L+L+DV + + L
Sbjct: 784 YVKKHGLVHLQQMLLSAIVGHNNSTMFENVDERVWKIKHMLNQKKVFLVLEDVHDSEVLK 963
Query: 315 ALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPS 374
A+ +FG GS+VIIT R K L HGI+ + VE +N+TEA +LL AF S +
Sbjct: 964 AIVKLSTFFGSGSKVIITAREKCFLEFHGIKRIYEVERMNKTEAFQLLNLKAFDSMNISP 1143
Query: 375 GYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYDA 434
+ IL YA G P +LE++GS L GKS+E+ + L Y I N++I+KIL+VS+DA
Sbjct: 1144CHVTILEGLETYASGHPFILEMIGSYLSGKSMEECESALHQYKEISNRDIKKILQVSFDA 1323
Query: 435 LEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGSI 494
LE+ +Q++ + IA + + + E++L Y C + + VL KSL+KI+
Sbjct: 1324LEKSQQNMLIHIALHLREQELEMVENLLHRKYGVCPKYDIRVLLNKSLIKIN------EN 1485
Query: 495 NDVRLHDLIKDMGKEVVRQESPKE 518
V +H L +DM VR + P E
Sbjct: 1486GHVIVHVLTQDM----VRDDIPVE 1545
Score = 145 bits (367), Expect(2) = 2e-85
Identities = 82/163 (50%), Positives = 104/163 (63%), Gaps = 1/163 (0%)
Frame = +3
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
MQ+PS ++Y F+SFRGSDTR GF+G L K L D+G TF D L+RG +IT
Sbjct: 120 MQTPSGLLGPK-AFRYSTFISFRGSDTRCGFSGFLNKYLIDRGFRTFFDDGELERGTQIT 296
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKT-KGRLVLPVFFGVEPTVVRH 119
+ KAIEESRIFIPV S NYASSSFCLDELV I+ +K GR V PVF+ + + V++
Sbjct: 297 VEIPKAIEESRIFIPVLSENYASSSFCLDELVKILEEFKKGNGRWVFPVFYYINISDVKN 476
Query: 120 RKGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH 162
+ GSYG+ALA H+ R ER + W AL+ A+ G H
Sbjct: 477 QTGSYGQALAVHKNRVM-----PERFEKWINALASVADFRGCH 590
>TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (13%)
Length = 581
Score = 313 bits (801), Expect = 3e-85
Identities = 158/189 (83%), Positives = 165/189 (86%)
Frame = +2
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
MQ PSSS+S S + YQVFLSFRG+DTR+GFTGNLYKALTDKGI TFID N LQRGDEIT
Sbjct: 17 MQLPSSSSSFSSAFTYQVFLSFRGTDTRHGFTGNLYKALTDKGIKTFIDDNDLQRGDEIT 196
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHR 120
PSLLKAIEESRIFIPVFSINYA+S FCLDELVHIIHCYKTKGRLVLPVFFGV+PT VRH
Sbjct: 197 PSLLKAIEESRIFIPVFSINYATSKFCLDELVHIIHCYKTKGRLVLPVFFGVDPTNVRHH 376
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYI 180
G YGEALA HEKRFQND NMERL WK AL+QAANLSGYH S PGYEYK IG IVKYI
Sbjct: 377 TGPYGEALAGHEKRFQNDKDNMERLHQWKLALTQAANLSGYHSS-PGYEYKFIGDIVKYI 553
Query: 181 SNKISRQPL 189
SNKISRQ L
Sbjct: 554 SNKISRQHL 580
>BF636566 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (20%)
Length = 663
Score = 309 bits (792), Expect = 3e-84
Identities = 155/202 (76%), Positives = 176/202 (86%)
Frame = +3
Query: 233 LARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIK 292
LA+ IYNFVADQFEG CFLH VRENS N+LK+LQ++LLLKT L IKL SEGIP+IK
Sbjct: 3 LAKAIYNFVADQFEGVCFLHKVRENSTHNSLKHLQKELLLKTVKLNIKLGDASEGIPLIK 182
Query: 293 ERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEG 352
ERL R KILLILDDVD L+QL ALAGGLDWFG GSRVIITTR+K LL+ HGIE T+AV G
Sbjct: 183 ERLNRMKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLTCHGIERTYAVNG 362
Query: 353 LNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHT 412
L+ETEA ELLRWMAFK+ +VPS Y D+LNRAVAYA GLPLVLE+VGSNLFGKS+E+W+ T
Sbjct: 363 LHETEAFELLRWMAFKNGEVPSSYNDVLNRAVAYASGLPLVLEIVGSNLFGKSMEEWQCT 542
Query: 413 LDGYDRIPNKEIQKILKVSYDA 434
LDGY++IPNKEIQ+ILKVSYDA
Sbjct: 543 LDGYEKIPNKEIQRILKVSYDA 608
Score = 42.0 bits (97), Expect = 0.001
Identities = 22/38 (57%), Positives = 25/38 (64%)
Frame = +1
Query: 415 GYDRIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKG 452
G R P K + LK + LEEE+QSVFLDIA CFKG
Sbjct: 550 GMKRFPIKRSKGYLK*AMMLLEEEQQSVFLDIAXCFKG 663
>BG646177 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 708
Score = 305 bits (782), Expect = 5e-83
Identities = 153/232 (65%), Positives = 186/232 (79%)
Frame = +3
Query: 254 VRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQL 313
VRE SA++ L+ LQEKLL KT GL +K HVSEGIP+IKERL KK+LLILDDVD LKQL
Sbjct: 3 VREISAKHGLEDLQEKLLSKTVGLSVKFGHVSEGIPIIKERLRLKKVLLILDDVDELKQL 182
Query: 314 HALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVP 373
LAG +W G GSRV++TTR+K LL+ HGIE T+ ++GLN+ EALELL+W AFK++KV
Sbjct: 183 KVLAGDPNWLGHGSRVVVTTRDKHLLACHGIERTYELDGLNKEEALELLKWKAFKNNKVD 362
Query: 374 SGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYD 433
S YE ILNRAV YA GLPL LEVVGS+LFGK ++WK TLD Y+RIP+KE+ KILKVS+D
Sbjct: 363 SSYEHILNRAVTYASGLPLALEVVGSSLFGKHKDEWKSTLDRYERIPHKEVLKILKVSFD 542
Query: 434 ALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKI 485
+LE++EQSVFLDIACCF+GY E EDIL AHY C+ +H+ VL K L+KI
Sbjct: 543 SLEKDEQSVFLDIACCFRGYILAEVEDILYAHYGECMKYHIRVLIEKCLIKI 698
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 302 bits (773), Expect = 5e-82
Identities = 159/222 (71%), Positives = 175/222 (78%), Gaps = 24/222 (10%)
Frame = +3
Query: 7 STSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKA 66
S+SISY +KYQVFLSFRGSDTRYGFTGNLYKALTDKGI+TFID + LQRGDEITPSL A
Sbjct: 3 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 182
Query: 67 IEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGE 126
IEESRIFIPVFS NYASSSFCLDELVHIIH YK GRLVLPVFFGV+P+ VRH +GSYGE
Sbjct: 183 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 362
Query: 127 ALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGK----------- 175
ALA+HE+RFQ++ +MERLQ WK AL+QAANLSG H S PGYEYKL GK
Sbjct: 363 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRS-PGYEYKLTGKIAFNQTPHLSS 539
Query: 176 -------------IVKYISNKISRQPLHVATYPVGLQSRVQQ 204
IVKYISNKI+R PLHVA YPVG + R+QQ
Sbjct: 540 DCSQRYEYDFIGDIVKYISNKINRVPLHVANYPVGFKFRIQQ 665
>TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance protein LM12
{Glycine max}, partial (25%)
Length = 677
Score = 293 bits (750), Expect = 2e-79
Identities = 149/205 (72%), Positives = 171/205 (82%)
Frame = +3
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
MQS SSS+SIS +KY VFLSFR DT YGFTGNLYKAL DKGI TFID N L+RGDE T
Sbjct: 66 MQSLSSSSSISSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDEST 245
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHR 120
PSL+KAIEESRI IP+FS NYASSSFCLDELVHIIHCYKT+G VLPVF+G +PT VRH+
Sbjct: 246 PSLVKAIEESRILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQ 425
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYI 180
GSYGE L +HE +FQN+ +NMERL+ WK AL+QAAN SG+H S GYEY+LI IV++I
Sbjct: 426 TGSYGEHLTKHEDKFQNNKENMERLKKWKMALTQAANFSGHHFS-QGYEYELIENIVEHI 602
Query: 181 SNKISRQPLHVATYPVGLQSRVQQV 205
S++I+R LHVA YPVGLQSRVQQV
Sbjct: 603 SDRINRVFLHVAKYPVGLQSRVQQV 677
>TC92005 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (7%)
Length = 858
Score = 290 bits (741), Expect = 3e-78
Identities = 156/236 (66%), Positives = 181/236 (76%), Gaps = 10/236 (4%)
Frame = +1
Query: 641 KVLTLDYCEYLTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEH 700
KVLTLD CEYLTHIPDVSGLSN+EK SF C NLITI +SIGH NKLE++SA GC KL+
Sbjct: 1 KVLTLDDCEYLTHIPDVSGLSNIEKFSFKFCRNLITIDDSIGHQNKLEFISAIGCSKLKR 180
Query: 701 FRPLGLASLKKLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELT 760
F PLGLASLK+L L C L++FPELLCKM +IK I NTSIGELP SFQNLSEL++++
Sbjct: 181 FPPLGLASLKELELSFCVSLNSFPELLCKMTNIKRILFVNTSIGELPSSFQNLSELNDIS 360
Query: 761 V--TSGMKFPK-------IVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFS-NF 810
+ ++FPK IVFSN+T+LSL NLSDECLPI+LKW VN+ LDLS + NF
Sbjct: 361 IERCGMLRFPKHNDKINSIVFSNVTQLSLQNCNLSDECLPILLKWFVNVKRLDLSHNFNF 540
Query: 811 KILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQ 866
ILPECL ECH + CC+SLEEIRGIPPNL+EL A C+SLSSSSRRML SQ
Sbjct: 541 NILPECLNECHLMKIFEFDCCKSLEEIRGIPPNLEELSAYKCESLSSSSRRMLTSQ 708
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 276 bits (705), Expect = 4e-74
Identities = 157/410 (38%), Positives = 249/410 (60%), Gaps = 4/410 (0%)
Frame = +3
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIP 75
Y VF++FRG DTR FT L+ AL KGI F D L +G+ I P LL+ IE S++F+
Sbjct: 69 YDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVA 248
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRF 135
V S NYASS++CL EL I C K G+ VLP+F+GV+P+ V+ + G Y + A+HE+RF
Sbjct: 249 VLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAKHEQRF 428
Query: 136 QNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKISRQPLHVATYP 195
+ DP + R W++AL+Q +++G+ D + + KIV+ I N + + V+
Sbjct: 429 KQDPHKVSR---WREALNQVGSIAGW-DLRDKQQSVEVEKIVQTILNILKCKSSFVSKDL 596
Query: 196 VGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVR 255
VG+ SR + +K L S DGV ++GI+G+GG GK+TLA +Y + +F+ SCF+ DV
Sbjct: 597 VGINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVS 776
Query: 256 ENSAQNNLKY-LQEKLLLKTTGLEIK--LDHVSEGIPVIKERLCRKKILLILDDVDNLKQ 312
+ ++ Q+++L +T G+E +H S +I+ RL R+K LLILD+VD ++Q
Sbjct: 777 KIFRLHDGPIDAQKQILHQTLGIEHHQICNHYS-ATDLIRHRLSREKTLLILDNVDQVEQ 953
Query: 313 LHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDK- 371
L + +W G GSR++I +R++ +L + ++ + V L+ TE+ +L R AFK +K
Sbjct: 954 LERIGVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKLEKI 1133
Query: 372 VPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPN 421
+ Y+++ + YA GLPL + V+GS L G+++ +WK L + PN
Sbjct: 1134IMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>AW686165 weakly similar to GP|18033111|gb functional candidate resistance
protein KR1 {Glycine max}, partial (7%)
Length = 551
Score = 274 bits (700), Expect = 1e-73
Identities = 135/174 (77%), Positives = 152/174 (86%)
Frame = +2
Query: 67 IEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGE 126
IEESRIFIPVFSINYASS FCLDELVHIIHCYKTKGRLVLPVF+GV+PT +RH+ GSYGE
Sbjct: 2 IEESRIFIPVFSINYASSKFCLDELVHIIHCYKTKGRLVLPVFYGVDPTQIRHQSGSYGE 181
Query: 127 ALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKISR 186
L +HE+ FQN+ KN ERL WK AL+QAANLSGYH S PGYEYK IGKIV+ ISNKI+R
Sbjct: 182 HLTKHEESFQNNKKNKERLHQWKLALTQAANLSGYHYS-PGYEYKFIGKIVEDISNKINR 358
Query: 187 QPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNF 240
LHVA YPVGL+SR++QVK LLD+ SD+GVHMVG+YG GGLGKSTLA+ IYNF
Sbjct: 359 VILHVAKYPVGLESRLEQVKLLLDKESDEGVHMVGLYGTGGLGKSTLAKAIYNF 520
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,171,289
Number of Sequences: 36976
Number of extensions: 577476
Number of successful extensions: 3700
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 3358
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3468
length of query: 1040
length of database: 9,014,727
effective HSP length: 106
effective length of query: 934
effective length of database: 5,095,271
effective search space: 4758983114
effective search space used: 4758983114
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC126790.9


