

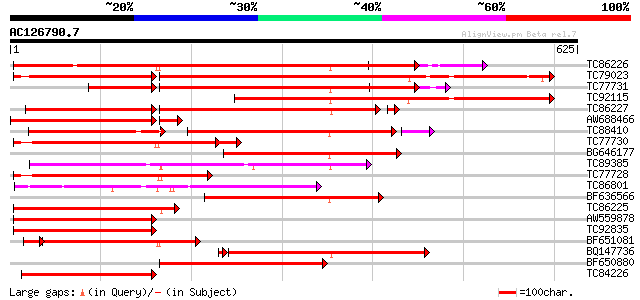
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.7 - phase: 0 /pseudo
(625 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 502 e-142
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 346 2e-95
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 335 3e-92
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 288 4e-78
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 275 4e-75
AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glyc... 258 9e-75
TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 174 2e-73
TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 239 7e-69
BG646177 weakly similar to GP|18033111|g functional candidate re... 254 5e-68
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 244 5e-65
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 242 4e-64
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 239 2e-63
BF636566 weakly similar to GP|9965103|gb| resistance protein LM6... 229 3e-60
TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance p... 224 1e-58
AW559878 weakly similar to GP|18033111|g functional candidate re... 223 1e-58
TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 217 1e-56
BF651081 weakly similar to GP|18033111|g functional candidate re... 203 1e-56
BQ147736 weakly similar to GP|18033111|g functional candidate re... 215 3e-56
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 213 2e-55
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 210 1e-54
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 502 bits (1292), Expect = e-142
Identities = 272/479 (56%), Positives = 335/479 (69%), Gaps = 32/479 (6%)
Frame = +3
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDD--KELPT 62
LMAMQLP SSS SY F++DVF+ +GTDTR+GFTGNL KAL DKGI TF DD +L
Sbjct: 36 LMAMQLPFYSSSFSYAFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQR 215
Query: 63 GDEITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPS 122
D++TP + IEESRI I IFS NYA+SS CLD LVHIIHC++ K V+PVF+G EP+
Sbjct: 216 RDKVTPII---IEESRILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPT 386
Query: 123 HVRKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALH------QFH------------- 163
VR YG+ALA+HE FQND +NMERL +WK AL +H
Sbjct: 387 DVRHHTGRYGKALAEHENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGK 566
Query: 164 ------SWINRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAE 217
+ I+R LHVA Y VGL+SR+ +V SLLD G DGV+++GI G GG GK+TLA
Sbjct: 567 IVKYISNKISRQSLHVATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLAR 746
Query: 218 AVYNSIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRL 277
A+YN + +QFE CFL VRENS +SLK QE LLSK++ L +EGI IIK+RL
Sbjct: 747 AIYNFVADQFEGLCFLEQVRENSASNSLKRFQEMLLSKTLQLKIKLADVSEGISIIKERL 926
Query: 278 CRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNK 337
CRKK+LLILDDVD QL L G WFG GSRVIITTRD++LL+CH I K Y LN
Sbjct: 927 CRKKILLILDDVDNMKQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNV 1106
Query: 338 EESLELLRKMTFKND---SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDK 394
E+LELLR M FKND SSY+ ILNR V YASGLP+ +++VGSNLFGK+I +C++TLD
Sbjct: 1107TEALELLRWMAFKNDKVPSSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDW 1286
Query: 395 YERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQR--HFNFIMAIQ*HI 451
YE+IP ++IQ+ILKVS+D+LEEE+QSVFLDIACCFKGC W+K + H ++ I H+
Sbjct: 1287YEKIPNKEIQRILKVSYDSLEEEEQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHV 1463
Score = 55.8 bits (133), Expect = 5e-08
Identities = 45/131 (34%), Positives = 68/131 (51%)
Frame = +1
Query: 396 ERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIMAIQ*HIMLEC 455
+R P + ++ LK + + + F + K + ++ +++F IM I * IML+C
Sbjct: 1291 KRFPIKRSKEYLK*AMILWRKRSKVSF*TLLVASKVVNGKRSKKYFMLIMVIA*IIMLKC 1470
Query: 456 WLMNLS*RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*N 515
L N+S L I T+ ++ HY+T** WV SD N L SLE G G+ I L F*
Sbjct: 1471 *LKNVS----LTILNTI-VMCHYIT**KTWVKNSSD*NHLLSLEKEAGYGLKRIYLKF*K 1635
Query: 516 KTHKIEMIYLN 526
KT + +I N
Sbjct: 1636 KTL*VRLICRN 1668
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 346 bits (887), Expect = 2e-95
Identities = 212/446 (47%), Positives = 284/446 (63%), Gaps = 11/446 (2%)
Frame = +3
Query: 166 INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN 225
I+R L VA+Y VGL+SR+ V LD D V+++G+ GTGG+GK+TLA+A+YN I +
Sbjct: 738 ISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIAD 917
Query: 226 QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
QFE CFL NVR NS +LK+LQE+LL K++ D L ++GI IIKQRLCRKK+LLI
Sbjct: 918 QFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLI 1097
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVDK +QLE L G WFG GSRVIITTR+++LL HGI + + LN E+LELLR
Sbjct: 1098 LDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLR 1277
Query: 346 KMTFKND--SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDI 403
M FK + SS++ ILNRA+ YASGLPLA+ ++GSNL G+S+ D STLD YE IP ++I
Sbjct: 1278 WMAFKENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEI 1457
Query: 404 QKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQR----HFNFIMAIQ*HIMLECWLMN 459
Q+ILKVS+D+LE+E+QSVFLDIACCFKGC W + + H+ + ++ E LM+
Sbjct: 1458 QRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMD 1637
Query: 460 LS*RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NK-TH 518
L + TL L+ M + + S L ++ ++ N T
Sbjct: 1638 ---HLKYDSYVTLHDLIEDMGKEVVR----QESPDEPGERSRLWFERDIVHVLKKNTGTR 1796
Query: 519 KIEMIYLNCSSME-PININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTS 577
KI+MI + SME I+ N AF+KM LKT I E G+ SK L+YLP SL V+ KG
Sbjct: 1797 KIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVM--KGCIP 1970
Query: 578 EPLSFCFS---FKASKIIVFSNCVYL 600
+ S S F+ K+++ +NC YL
Sbjct: 1971 KSPSSSSSNKKFEDMKVLILNNCEYL 2048
Score = 195 bits (495), Expect = 5e-50
Identities = 98/157 (62%), Positives = 118/157 (74%)
Frame = +1
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGD 64
LMA Q P+S F + VF+SFRG DTR GFTGNLYKAL+DKGI+TFIDD +L GD
Sbjct: 61 LMATQSPSS-------FTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGD 219
Query: 65 EITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHV 124
EITPSL+ +IE+SRI I +FS+NYA+SSFCLDELVHI HC+ K V+PVF G +P+ V
Sbjct: 220 EITPSLKNAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDV 399
Query: 125 RKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ 161
R YGEALA H+ +FQND +N ERL +WKEAL Q
Sbjct: 400 RHHTGRYGEALAVHKKKFQNDKDNTERLQQWKEALSQ 510
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 335 bits (859), Expect = 3e-92
Identities = 169/291 (58%), Positives = 221/291 (75%), Gaps = 5/291 (1%)
Frame = +1
Query: 166 INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN 225
IN L+VA+Y VGL+SRI +V LLD+G D V ++G+ GTGG+GK+TLA+AVYN + +
Sbjct: 547 INHVFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVYNFVAD 726
Query: 226 QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
QFE CFL+NVRENS +LK+LQ++LLSK + +D LE +EGI IIK+RL RKK+LLI
Sbjct: 727 QFEGVCFLHNVRENSTHKNLKHLQKKLLSKIVKFDGKLEDVSEGIPIIKERLSRKKILLI 906
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVDK QLE L G WFG GSRVIITTRD++LL+CHGIT + + LN+ E+LELLR
Sbjct: 907 LDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLACHGITSTHAVEELNETEALELLR 1086
Query: 346 KMTFKND---SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPED 402
+M FKND S+Y+ ILNR V YASGLPLA+ +G NLFG+ + D + LD+YE IP +D
Sbjct: 1087RMAFKNDKVPSTYEEILNRVVTYASGLPLAIVTIGDNLFGRKVEDWKRILDEYENIPNKD 1266
Query: 403 IQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQR--HFNFIMAIQ*HI 451
IQ+IL+VS+D LE +++SVFLDIACCFKGC W K ++ H ++ I+ H+
Sbjct: 1267IQRILQVSYDALEPKEKSVFLDIACCFKGCKWTKVKKILHAHYGHCIEHHV 1419
Score = 102 bits (254), Expect = 4e-22
Identities = 47/74 (63%), Positives = 56/74 (75%)
Frame = +2
Query: 88 YATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEFQNDME 147
YA+SSFCLDELVHIIHC++ K V+PVFY EP+H+R SYGE L KHE FQN+ +
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 148 NMERLLKWKEALHQ 161
NMERL +WK AL Q
Sbjct: 182 NMERLRQWKIALTQ 223
Score = 47.0 bits (110), Expect = 2e-05
Identities = 33/91 (36%), Positives = 49/91 (53%), Gaps = 1/91 (1%)
Frame = +2
Query: 397 RIPPEDIQK-ILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIMAIQ*HIMLEC 455
+I P I K K++ + ++ F + KG + Q+ +++F IMAI *+IMLEC
Sbjct: 1247 KIFPIKISKGYFKLAMMLWNQRRRVSF*TLLVASKGVNGQRLKKYFMLIMAIA*NIMLEC 1426
Query: 456 WLMNLS*RLVLQIHTTLLILLHYMT**SIWV 486
WL NLS* + + + YMT** WV
Sbjct: 1427 WLKNLS*AI-----GNMTLR*PYMT**RTWV 1504
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 288 bits (737), Expect = 4e-78
Identities = 181/363 (49%), Positives = 230/363 (62%), Gaps = 10/363 (2%)
Frame = +2
Query: 248 LQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQ 307
LQE+LL K++ L +EGI IK+RL KK LLILDDVD QL L G P WFG+
Sbjct: 2 LQEELLLKALQLKIKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFGR 181
Query: 308 GSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKND---SSYDYILNRAVE 364
GSRVIITTRD++LL HGI +E + L E+LELLR M FKN+ S Y+ +LNRAV
Sbjct: 182 GSRVIITTRDKHLLRSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSIYEDVLNRAVS 361
Query: 365 YASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTLEEEQQSVFLD 424
YASGLPL L++VGSNLFGK+I + + TLD YE+IP + I +ILKVS+D LEEEQQSVFLD
Sbjct: 362 YASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEEQQSVFLD 541
Query: 425 IACCFKGCDWQKFQ----RHFNFIMAIQ*HIMLECWLMNLS*RLVLQIH-TTLLILLHYM 479
IACCFKGC W++F+ H+ + ++ E L+ ++ I+ TL L+ M
Sbjct: 542 IACCFKGCGWEEFEYILRAHYGHRITHHLVVLAEKSLVKITHPHYGSIYELTLHDLIKEM 721
Query: 480 T**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NK-THKIEMIYLNCSSME-PININE 537
+ + + S L C ++ ++ N T KIEMIY+N S E I+
Sbjct: 722 GKEVVR----QESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNFPSEEFVIDKKG 889
Query: 538 KAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSEPLSFCFSFKASKIIVFSNC 597
KAFKKM +LKTLIIE +FSKGLKYLP SL VLK +G SE L C KA +I FS C
Sbjct: 890 KAFKKMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRGCLSESLISCSLSKAIEITSFSYC 1069
Query: 598 VYL 600
+YL
Sbjct: 1070IYL 1078
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 275 bits (703), Expect(2) = 4e-75
Identities = 142/246 (57%), Positives = 187/246 (75%), Gaps = 3/246 (1%)
Frame = +1
Query: 166 INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN 225
INR LHVA+Y VGL+SR+ +V LLD +GV ++G+ GT LGK+TLA+A+YN I +
Sbjct: 730 INRVFLHVAKYPVGLQSRVQQVKLLLDEESDEGVNMVGLYGTRALGKSTLAKAIYNFIAD 909
Query: 226 QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
QFE CFL+NVRENS + +LK+LQ++LLSK++ + L +EGI IIK+RLCRKK+LLI
Sbjct: 910 QFEGVCFLHNVRENSARKNLKHLQKELLSKTVQLNIKLRDVSEGIPIIKERLCRKKILLI 1089
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVD+ +QLE L G WFG GSRVIITTRD++LL+CHGI + Y L +E+LELLR
Sbjct: 1090LDDVDQLDQLEALAGGLDWFGPGSRVIITTRDKHLLTCHGIERTYAVRGLYGKEALELLR 1269
Query: 346 KMTFKNDS---SYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPED 402
FKN+ SY+ +LNRAV Y SG+PL L++VGSNLFGK+I ++TLD Y+RIP ++
Sbjct: 1270WTAFKNNKVPPSYEDVLNRAVSYGSGIPLVLEIVGSNLFGKNIEVWKNTLDGYDRIPNKE 1449
Query: 403 IQKILK 408
IQKIL+
Sbjct: 1450IQKILR 1467
Score = 196 bits (498), Expect = 2e-50
Identities = 97/144 (67%), Positives = 111/144 (76%)
Frame = +3
Query: 18 SYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEES 77
S F + VF+SFR DT +GFTGNLYKAL DKGI TFIDD +L GDE TPSL K+IEES
Sbjct: 3 SSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEES 182
Query: 78 RIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAK 137
RI I IFS NYA+SSFCLDELVHIIHC++ + V+PVFYG +P+HVR SYGE L K
Sbjct: 183 RILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTK 362
Query: 138 HEVEFQNDMENMERLLKWKEALHQ 161
HE +FQN+ ENMERL KWK AL Q
Sbjct: 363 HEDKFQNNKENMERLKKWKMALTQ 434
Score = 25.4 bits (54), Expect(2) = 4e-75
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = +2
Query: 417 EQQSVFLDIACCF 429
E+Q+VFLDIAC F
Sbjct: 1466 EEQNVFLDIACFF 1504
>AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glycine max},
partial (31%)
Length = 668
Score = 258 bits (660), Expect(2) = 9e-75
Identities = 130/160 (81%), Positives = 139/160 (86%)
Frame = +2
Query: 2 PFFLMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELP 61
PF LMAMQ P+SSSSLSY F+FDVFISFRGTDTR+GFTGNLYKALSDKGI TFIDDKEL
Sbjct: 23 PFLLMAMQSPSSSSSLSYGFSFDVFISFRGTDTRYGFTGNLYKALSDKGIRTFIDDKELQ 202
Query: 62 TGDEITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEP 121
GDEITPSL KSIE+SRIAII+FSK+YA+SSFCL ELVHII C EK T VIPVFYGTEP
Sbjct: 203 RGDEITPSLLKSIEDSRIAIIVFSKDYASSSFCLXELVHIIQCSNEKGTTVIPVFYGTEP 382
Query: 122 SHVRKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ 161
S VR DSYGEALAKHE FQN ENMERLLKWK+AL+Q
Sbjct: 383 SQVRHQNDSYGEALAKHEEGFQNSKENMERLLKWKKALNQ 502
Score = 40.8 bits (94), Expect(2) = 9e-75
Identities = 19/25 (76%), Positives = 22/25 (88%)
Frame = +3
Query: 166 INRCHLHVAEYLVGLESRISEVNSL 190
IN LHVA+YL+GL+SRISEVNSL
Sbjct: 594 INHVPLHVADYLIGLKSRISEVNSL 668
>TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (8%)
Length = 2252
Score = 174 bits (441), Expect(3) = 2e-73
Identities = 102/236 (43%), Positives = 145/236 (61%), Gaps = 6/236 (2%)
Frame = +1
Query: 197 DGVYIIGILGTGGLGKTTLAEAVYN-SIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSK 255
DGV ++GI G G+GKTTLA VY+ +F+ CF NV E KH L +LQ+ LLS
Sbjct: 655 DGVCLVGICGVPGIGKTTLARGVYHFGGGTEFDSCCFFDNVGEYVKKHGLVHLQQMLLSA 834
Query: 256 SIGYD--TPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVII 313
+G++ T E+ +E + IK L +KKV L+L+DV L+ +V +FG GS+VII
Sbjct: 835 IVGHNNSTMFENVDERVWKIKHMLNQKKVFLVLEDVHDSEVLKAIVKLSTFFGSGSKVII 1014
Query: 314 TTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---DSSYDYILNRAVEYASGLP 370
T R++ L HGI +IYE + +NK E+ +LL F + + IL YASG P
Sbjct: 1015TAREKCFLEFHGIKRIYEVERMNKTEAFQLLNLKAFDSMNISPCHVTILEGLETYASGHP 1194
Query: 371 LALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTLEEEQQSVFLDIA 426
L+++GS L GKS+ +CES L +Y+ I DI+KIL+VSFD LE+ QQ++ + IA
Sbjct: 1195FILEMIGSYLSGKSMEECESALHQYKEISNRDIKKILQVSFDALEKSQQNMLIHIA 1362
Score = 120 bits (302), Expect(3) = 2e-73
Identities = 72/152 (47%), Positives = 92/152 (60%), Gaps = 1/152 (0%)
Frame = +3
Query: 21 FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIA 80
F + FISFRG+DTR GF+G L K L D+G TF DD EL G +IT + K+IEESRI
Sbjct: 156 FRYSTFISFRGSDTRCGFSGFLNKYLIDRGFRTFFDDGELERGTQITVEIPKAIEESRIF 335
Query: 81 IIIFSKNYATSSFCLDELVHIIHCFREKVTK-VIPVFYGTEPSHVRKLEDSYGEALAKHE 139
I + S+NYA+SSFCLDELV I+ F++ + V PVFY S V+ SYG+ALA H+
Sbjct: 336 IPVLSENYASSSFCLDELVKILEEFKKGNGRWVFPVFYYINISDVKNQTGSYGQALAVHK 515
Query: 140 VEFQNDMENMERLLKWKEALHQFHSWINRCHL 171
+ ER KW AL + CH+
Sbjct: 516 -----NRVMPERFEKWINALASVADF-RGCHM 593
Score = 21.2 bits (43), Expect(3) = 2e-73
Identities = 12/36 (33%), Positives = 21/36 (58%)
Frame = +2
Query: 433 DWQKFQRHFNFIMAIQ*HIMLECWLMNLS*RLVLQI 468
+W+ + F M +++LE L+NLS*RL+ +
Sbjct: 1382 NWKWLKTCFIVNMECARNMILEFCLINLS*RLMRMV 1489
>TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (80%)
Length = 831
Score = 239 bits (610), Expect(2) = 7e-69
Identities = 133/253 (52%), Positives = 167/253 (65%), Gaps = 25/253 (9%)
Frame = +1
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGD 64
LMAMQ P+ VF++FRG+DTR FTGNLYKAL DKGI TFID+ +L GD
Sbjct: 13 LMAMQSPSR-----------VFLNFRGSDTRNNFTGNLYKALVDKGIRTFIDENDLQRGD 159
Query: 65 EITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHV 124
EITPSL K+IEESRI I IFS NYA+SSFCLDELVHIIHC++ K V+PVFYG +P+ V
Sbjct: 160 EITPSLVKAIEESRICIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYGVDPTDV 339
Query: 125 RKLEDSYGEALAKHEVEFQNDMENMERLLKWKEAL-----------------HQF----- 162
R+ SYGE L KHE FQN+ +NMERL +WK AL H+F
Sbjct: 340 RRHTCSYGEYLTKHEEGFQNNEKNMERLRQWKMALTQAANLSGYHYSPHEYEHKFIEKIV 519
Query: 163 ---HSWINRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAV 219
+ IN L+VA+Y VGL+SRI +V LLD+G D V ++G+ GTGG+GK+TLA+AV
Sbjct: 520 QYISNNINHDFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAV 699
Query: 220 YNSIVNQFECRCF 232
+N + + + F
Sbjct: 700 FNFLADHLKVYVF 738
Score = 40.4 bits (93), Expect(2) = 7e-69
Identities = 19/29 (65%), Positives = 24/29 (82%)
Frame = +3
Query: 227 FECRCFLYNVRENSFKHSLKYLQEQLLSK 255
FE CFL+NVRENS ++LK+LQ+ LLSK
Sbjct: 720 FEGVCFLHNVRENSSHNNLKHLQKMLLSK 806
>BG646177 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 708
Score = 254 bits (650), Expect = 5e-68
Identities = 129/199 (64%), Positives = 161/199 (80%), Gaps = 3/199 (1%)
Frame = +3
Query: 236 VRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQL 295
VRE S KH L+ LQE+LLSK++G H +EGI IIK+RL KKVLLILDDVD+ QL
Sbjct: 3 VREISAKHGLEDLQEKLLSKTVGLSVKFGHVSEGIPIIKERLRLKKVLLILDDVDELKQL 182
Query: 296 EKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---D 352
+ L G+P W G GSRV++TTRD++LL+CHGI + YE D LNKEE+LELL+ FKN D
Sbjct: 183 KVLAGDPNWLGHGSRVVVTTRDKHLLACHGIERTYELDGLNKEEALELLKWKAFKNNKVD 362
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
SSY++ILNRAV YASGLPLAL+VVGS+LFGK + +STLD+YERIP +++ KILKVSFD
Sbjct: 363 SSYEHILNRAVTYASGLPLALEVVGSSLFGKHKDEWKSTLDRYERIPHKEVLKILKVSFD 542
Query: 413 TLEEEQQSVFLDIACCFKG 431
+LE+++QSVFLDIACCF+G
Sbjct: 543 SLEKDEQSVFLDIACCFRG 599
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 244 bits (624), Expect = 5e-65
Identities = 149/411 (36%), Positives = 227/411 (54%), Gaps = 34/411 (8%)
Frame = +3
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF++FRG DTR FT L+ AL KGI+ F DD LP G+ I P L ++IE S++ +
Sbjct: 69 YDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVA 248
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ S+NYA+S++CL EL I C + V+P+FYG +PS V+K Y + AKHE F
Sbjct: 249 VLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAKHEQRF 428
Query: 143 QNDMENMERLLKWKEALHQFHS---W--------------------INRC-HLHVAEYLV 178
+ D + R W+EAL+Q S W I +C V++ LV
Sbjct: 429 KQDPHKVSR---WREALNQVGSIAGWDLRDKQQSVEVEKIVQTILNILKCKSSFVSKDLV 599
Query: 179 GLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNVRE 238
G+ SR + L L DGV +IGI G GG GKTTLA +Y I ++F+ CF+ +V +
Sbjct: 600 GINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVSK 779
Query: 239 NSFKHSLKY-LQEQLLSKSIGYDTPLEHDN-----EGIEIIKQRLCRKKVLLILDDVDKP 292
H Q+Q+L +++G +EH ++I+ RL R+K LLILD+VD+
Sbjct: 780 IFRLHDGPIDAQKQILHQTLG----IEHHQICNHYSATDLIRHRLSREKTLLILDNVDQV 947
Query: 293 NQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKND 352
QLE++ W G GSR++I +RD ++L + + Y+ L+ ES +L R+ FK +
Sbjct: 948 EQLERIGVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKLE 1127
Query: 353 ----SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIP 399
+Y + + YA+GLPL + V+GS L G+++ + +S L + + P
Sbjct: 1128KIIMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRP 1280
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 242 bits (617), Expect = 4e-64
Identities = 135/244 (55%), Positives = 160/244 (65%), Gaps = 25/244 (10%)
Frame = +1
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGD 64
LMAMQ P+ VF+SFRG+DTR FTGNLYKAL DKGI TFIDD +L GD
Sbjct: 67 LMAMQSPSR-----------VFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGD 213
Query: 65 EITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHV 124
EITPSL K+IEESRI I IFS NYA+SSFCLDELVHIIHC++ K V PVFY EP+H+
Sbjct: 214 EITPSLVKAIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHI 393
Query: 125 RKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQF----------HSW--------- 165
R YGE L KHE FQN+ +NMERL +WK AL Q H +
Sbjct: 394 RNQSGIYGEHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHGYEYKFIEKIV 573
Query: 166 ------INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAV 219
IN L+VA+Y VGL+SRI EV LLD+G D V ++G+ GTGG+GK+TLA+AV
Sbjct: 574 EDISNNINHVFLNVAKYPVGLQSRIEEVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAV 753
Query: 220 YNSI 223
YN +
Sbjct: 754 YNFV 765
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 239 bits (611), Expect = 2e-63
Identities = 152/367 (41%), Positives = 211/367 (57%), Gaps = 29/367 (7%)
Frame = +2
Query: 6 MAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDE 65
MAM L +SS S + +DVFISFRG DTR GFT +L+ ALS +HT+ID + + GDE
Sbjct: 20 MAMSLASSSHSAAQK-KYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYIDYR-IEKGDE 193
Query: 66 ITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTK---VIPVFYGTEPS 122
+ P L K+I++S + +++FS+NYA+S++CL+ELV ++ C + VIPVFY +PS
Sbjct: 194 VWPELEKAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPVFYHVDPS 373
Query: 123 HVRKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ------FHSWINRCHLHVAE- 175
HVRK SYG ALAKH+ E Q+D + + WK AL Q FHS R ++ E
Sbjct: 374 HVRKQTGSYGSALAKHKQENQDD----KMMQNWKNALFQAANLSGFHSSTYRTESNMIED 541
Query: 176 ---YLVG---------------LESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAE 217
L+G L+ V SL+ T V IIG+ G GG GKTTLA
Sbjct: 542 ITRALLGKLNHQYRDELTCNLILDENYWAVRSLIKFDSTT-VQIIGLWGMGGTGKTTLAA 718
Query: 218 AVYNSIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRL 277
A++ ++E CFL V E S KH + Y +LLSK +G D ++ +IK+RL
Sbjct: 719 AMFQRFSFKYEGNCFLERVTEVSKKHGINYTCNKLLSKLLGEDLRIDTPKVIPAMIKRRL 898
Query: 278 CRKKVLLILDDVDKPNQLEKLVG-EPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLN 336
K ++LDDV L+ L+G GW G GS VI+TTRD+++L GI +IYE +N
Sbjct: 899 RHMKSFIVLDDVHNSELLQDLIGVRGGWLGPGSIVIVTTRDKHVLISGGIDEIYEVKKMN 1078
Query: 337 KEESLEL 343
+ SL+L
Sbjct: 1079SQNSLQL 1099
>BF636566 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (20%)
Length = 663
Score = 229 bits (583), Expect = 3e-60
Identities = 118/201 (58%), Positives = 154/201 (75%), Gaps = 3/201 (1%)
Frame = +3
Query: 215 LAEAVYNSIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIK 274
LA+A+YN + +QFE CFL+ VRENS +SLK+LQ++LL K++ + L +EGI +IK
Sbjct: 3 LAKAIYNFVADQFEGVCFLHKVRENSTHNSLKHLQKELLLKTVKLNIKLGDASEGIPLIK 182
Query: 275 QRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADS 334
+RL R K+LLILDDVDK QLE L G WFG GSRVIITTRD++LL+CHGI + Y +
Sbjct: 183 ERLNRMKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLTCHGIERTYAVNG 362
Query: 335 LNKEESLELLRKMTFKN---DSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCEST 391
L++ E+ ELLR M FKN SSY+ +LNRAV YASGLPL L++VGSNLFGKS+ + + T
Sbjct: 363 LHETEAFELLRWMAFKNGEVPSSYNDVLNRAVAYASGLPLVLEIVGSNLFGKSMEEWQCT 542
Query: 392 LDKYERIPPEDIQKILKVSFD 412
LD YE+IP ++IQ+ILKVS+D
Sbjct: 543 LDGYEKIPNKEIQRILKVSYD 605
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/36 (58%), Positives = 25/36 (69%)
Frame = +1
Query: 396 ERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
+R P + + LK + LEEEQQSVFLDIA CFKG
Sbjct: 556 KRFPIKRSKGYLK*AMMLLEEEQQSVFLDIAXCFKG 663
>TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance protein LM12
{Glycine max}, partial (25%)
Length = 677
Score = 224 bits (570), Expect = 1e-58
Identities = 120/207 (57%), Positives = 142/207 (67%), Gaps = 24/207 (11%)
Frame = +3
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGD 64
LMAMQ +SSSS+S F + VF+SFR DT +GFTGNLYKAL DKGI TFIDD +L GD
Sbjct: 57 LMAMQSLSSSSSISSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGD 236
Query: 65 EITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHV 124
E TPSL K+IEESRI I IFS NYA+SSFCLDELVHIIHC++ + V+PVFYG +P+HV
Sbjct: 237 ESTPSLVKAIEESRILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHV 416
Query: 125 RKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQFHSW------------------- 165
R SYGE L KHE +FQN+ ENMERL KWK AL Q ++
Sbjct: 417 RHQTGSYGEHLTKHEDKFQNNKENMERLKKWKMALTQAANFSGHHFSQGYEYELIENIVE 596
Query: 166 -----INRCHLHVAEYLVGLESRISEV 187
INR LHVA+Y VGL+SR+ +V
Sbjct: 597 HISDRINRVFLHVAKYPVGLQSRVQQV 677
>AW559878 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (12%)
Length = 586
Score = 223 bits (569), Expect = 1e-58
Identities = 116/159 (72%), Positives = 130/159 (80%), Gaps = 2/159 (1%)
Frame = +2
Query: 5 LMAMQLPTSSSS--LSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPT 62
LM MQLP+SSSS LS DF +DVFISFRG DTR GFTG+LYKAL DKGI TFIDDKEL
Sbjct: 41 LMVMQLPSSSSSSSLSNDFIYDVFISFRGIDTRSGFTGHLYKALCDKGIRTFIDDKELQR 220
Query: 63 GDEITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPS 122
GDEITPSL KSIE S+IAII+FS+NYATSSFCLDELVHII+ F+EK V+PVFYG EPS
Sbjct: 221 GDEITPSLLKSIEHSKIAIIVFSENYATSSFCLDELVHIINYFKEKGRLVLPVFYGVEPS 400
Query: 123 HVRKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ 161
HVR + YGEAL + E FQN+ ENM+RL KWK AL+Q
Sbjct: 401 HVRHQNNKYGEALTEFEEMFQNNKENMDRLQKWKIALNQ 517
>TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (13%)
Length = 581
Score = 217 bits (552), Expect = 1e-56
Identities = 108/157 (68%), Positives = 125/157 (78%)
Frame = +2
Query: 5 LMAMQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGD 64
+MAMQLP+SSSS S F + VF+SFRGTDTR GFTGNLYKAL+DKGI TFIDD +L GD
Sbjct: 8 IMAMQLPSSSSSFSSAFTYQVFLSFRGTDTRHGFTGNLYKALTDKGIKTFIDDNDLQRGD 187
Query: 65 EITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHV 124
EITPSL K+IEESRI I +FS NYATS FCLDELVHIIHC++ K V+PVF+G +P++V
Sbjct: 188 EITPSLLKAIEESRIFIPVFSINYATSKFCLDELVHIIHCYKTKGRLVLPVFFGVDPTNV 367
Query: 125 RKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ 161
R YGEALA HE FQND +NMERL +WK AL Q
Sbjct: 368 RHHTGPYGEALAGHEKRFQNDKDNMERLHQWKLALTQ 478
>BF651081 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (16%)
Length = 664
Score = 203 bits (516), Expect(2) = 1e-56
Identities = 111/199 (55%), Positives = 134/199 (66%), Gaps = 25/199 (12%)
Frame = +1
Query: 37 GFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAIIIFSKNYATSSFCLD 96
GFTGNLYKAL DKGIHTFID+ EL GDEITPSL K+IEESRI I +FS NYA+SSFCLD
Sbjct: 67 GFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYASSSFCLD 246
Query: 97 ELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEFQNDMENMERLLKWK 156
ELVHIIHC++ K V+PVF+ EP+ VR + SYGEALA+HE FQND ++MERL WK
Sbjct: 247 ELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSMERLQGWK 426
Query: 157 EALHQ------FH-------------------SWINRCHLHVAEYLVGLESRISEVNSLL 191
EAL Q +H + I++ LHVA Y VGL+S ++ SLL
Sbjct: 427 EALSQAANLSGYHDSPPGYEYKLIGKIVKYIXNKISQQPLHVATYPVGLQSXXQQMKSLL 606
Query: 192 DLGCTDGVYIIGILGTGGL 210
D GV+++GI G GGL
Sbjct: 607 DEXSDHGVHMVGIYGIGGL 663
Score = 35.4 bits (80), Expect(2) = 1e-56
Identities = 12/23 (52%), Positives = 20/23 (86%)
Frame = +3
Query: 16 SLSYDFNFDVFISFRGTDTRFGF 38
S+SY++ + VF++FRG+DTR+ F
Sbjct: 3 SISYEYKYQVFLNFRGSDTRYXF 71
>BQ147736 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 712
Score = 215 bits (547), Expect(2) = 3e-56
Identities = 122/224 (54%), Positives = 155/224 (68%), Gaps = 3/224 (1%)
Frame = +2
Query: 242 KHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGE 301
K+ L++LQ +LLSK + D L NEGI IIKQRL RKKVLLILDDV + QL+ L G
Sbjct: 41 KYGLEHLQGKLLSKLVELDIKLVDVNEGIPIIKQRLHRKKVLLILDDVHELKQLQVLAGG 220
Query: 302 PGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKNDS---SYDYI 358
WFG GS+VIITTRD LL+ HGI + YE D LN++E+LELLR FKN+ ++D +
Sbjct: 221 LDWFGPGSKVIITTRDEQLLAGHGIERAYEIDKLNEKEALELLRWNAFKNNKVNPNFDNV 400
Query: 359 LNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTLEEEQ 418
L+ AV YASGLPLAL+VVGSNLFGK+I + +S L++YERIP + IQ+ILKVSFD L E+
Sbjct: 401 LHLAVTYASGLPLALEVVGSNLFGKNIGEWKSALNQYERIPIKKIQEILKVSFDALXEDX 580
Query: 419 QSVFLDIACCFKGCDWQKFQRHFNFIMAIQ*HIMLECWLMNLS* 462
+VFLDI+C K CD + + + L C + NLS*
Sbjct: 581 XNVFLDISCFLKXCDLKXVEDILHAHYGSCXKHQLXCCMKNLS* 712
Score = 22.3 bits (46), Expect(2) = 3e-56
Identities = 8/10 (80%), Positives = 10/10 (100%)
Frame = +1
Query: 231 CFLYNVRENS 240
CFL++VRENS
Sbjct: 7 CFLHDVRENS 36
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 213 bits (542), Expect = 2e-55
Identities = 109/185 (58%), Positives = 135/185 (72%)
Frame = +3
Query: 166 INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN 225
INR LHVA Y VGL SR+ EV SLLD G DGV+++GI G GGLGK+ LA A+YN + +
Sbjct: 75 INRQPLHVANYPVGLHSRVQEVKSLLDEGPDDGVHMVGIYGIGGLGKSALARAIYNFVAD 254
Query: 226 QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
QFE CFL++VRENS +++LK+LQE+LL K+ G L+H EGI IIK+RLCR K+LLI
Sbjct: 255 QFEGLCFLHDVRENSAQNNLKHLQEKLLLKTTGLKIKLDHVCEGIPIIKERLCRNKILLI 434
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVD QL L G P WFG GSRVIITTRD++LL+ H I + Y + L ++LELLR
Sbjct: 435 LDDVDDMEQLHALAGGPDWFGHGSRVIITTRDKHLLTSHDIERTYAVEGLYGTKALELLR 614
Query: 346 KMTFK 350
M FK
Sbjct: 615 XMAFK 629
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 210 bits (535), Expect = 1e-54
Identities = 102/148 (68%), Positives = 121/148 (80%)
Frame = +3
Query: 14 SSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKS 73
SSS+SY F + VF+SFRG+DTR+GFTGNLYKAL+DKGIHTFIDD EL GDEITPSL +
Sbjct: 3 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 182
Query: 74 IEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGE 133
IEESRI I +FS NYA+SSFCLDELVHIIH +++ V+PVF+G +PSHVR SYGE
Sbjct: 183 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 362
Query: 134 ALAKHEVEFQNDMENMERLLKWKEALHQ 161
ALAKHE FQ++ ++MERL KWK AL Q
Sbjct: 363 ALAKHEERFQHNTDHMERLQKWKIALTQ 446
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,795,095
Number of Sequences: 36976
Number of extensions: 305243
Number of successful extensions: 2403
Number of sequences better than 10.0: 235
Number of HSP's better than 10.0 without gapping: 2240
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2282
length of query: 625
length of database: 9,014,727
effective HSP length: 102
effective length of query: 523
effective length of database: 5,243,175
effective search space: 2742180525
effective search space used: 2742180525
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126790.7


