

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.10 + phase: 0
(1621 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
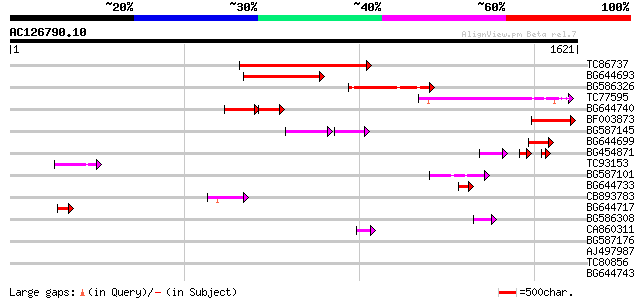
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 294 2e-79
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 254 2e-67
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 224 2e-58
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 196 8e-50
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 89 1e-32
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 130 4e-30
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 94 2e-29
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 90 8e-18
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 66 1e-17
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 74 6e-13
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 64 6e-10
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 53 8e-07
CB893783 weakly similar to GP|22830935|dbj hypothetical protein~... 48 4e-05
BG644717 46 1e-04
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 43 9e-04
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 43 9e-04
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 43 0.001
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 42 0.001
TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protei... 39 0.013
BG644743 39 0.013
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 294 bits (752), Expect = 2e-79
Identities = 170/392 (43%), Positives = 238/392 (60%), Gaps = 14/392 (3%)
Frame = +1
Query: 656 EFSDVF-PEELPGIPPDREI-EFSIDLIP---GTQPISI--PPYRMAPAELKELREQLQD 708
EF D+F PE+ +P R + + +I LIP G P P Y M+ EL L++ L+D
Sbjct: 352 EFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKTLED 531
Query: 709 LLDKGFIRASTSPWGAPVLFVKKKDGSMRLCVDYRQLNKVTIKNKYPLPRIDELFDQLQG 768
LLDKGFI+AS S GAPVLFV+K G +R CVDYR LN +T K++YPLP I E ++ G
Sbjct: 532 LLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAG 711
Query: 769 AQCFSKIDLRSGYHQLKIKSEDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFK 828
A+ F+K+D+ + +H+++IK ED KTAFRTRYG +E++V FGLT APA F +N+
Sbjct: 712 ARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLH 891
Query: 829 PFLDRFVIVFIDDILIYSK-SREEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGH 887
FLD FV +IDD+LIY+ S+++HE +R VL+ L + L KCEF + +V ++G
Sbjct: 892 EFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGF 1071
Query: 888 IVSK-NGISVDPSKVEAVQNWPRPTSVKEIRSFLGLAGYYRRFVKDFSKLAFPLTRLTQK 946
I++ G+S DP K+ A+++W P SVK RSFLG YY+ F+ +S++ PLTRLT+K
Sbjct: 1072 ILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRK 1251
Query: 947 KVEFKWTDACEESFQKLKECLISAPILALPTSGGGYVVYCDASRVGLGCVLMQH-----G 1001
F+W E +F KLK P+L + V D S LG VL Q
Sbjct: 1252 DFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGTGAA 1431
Query: 1002 KVIAYASRQLKRHEQNYPTHDLEMAAVIFALK 1033
+A+ S++L E NYP HD E+ AV L+
Sbjct: 1432 HPVAFHSQRLSPAEYNYPIHDKELLAVWACLR 1527
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 254 bits (648), Expect = 2e-67
Identities = 132/233 (56%), Positives = 166/233 (70%)
Frame = +2
Query: 668 IPPDREIEFSIDLIPGTQPISIPPYRMAPAELKELREQLQDLLDKGFIRASTSPWGAPVL 727
+PP+ +I+F IDL+P PI IP YR+ P +LK L+ QL+DLL+KGFI+ S P G VL
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 728 FVKKKDGSMRLCVDYRQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQLKIK 787
F+KKKDG +R+ +DY QLN V IK KYPLP IDELFD LQG++ F KIDLR G HQ ++
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 788 SEDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSK 847
ED+ KTAFR RYGHYE LVMSFG TN P AFM+LMNRVF+ +LD VIVF +DILIYSK
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 848 SREEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSK 900
+ EHE HLRL L+ L++ L + S +E F H++S G+ VD +
Sbjct: 557 NENEHENHLRLALKVLKDIGL-CQISYV*ILVEVGFFSLHVISGEGLKVDSKR 712
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 224 bits (571), Expect = 2e-58
Identities = 125/247 (50%), Positives = 158/247 (63%), Gaps = 1/247 (0%)
Frame = +2
Query: 969 SAPILALPTSGGGYVVYCDASRVGLGCVLMQHGKVIAYASRQLKRHEQNYPTHDLEMAAV 1028
SAPIL LP YVVY DAS GLGCVL QH KVIAYASRQL++HE NYPTHDLEMAAV
Sbjct: 11 SAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAAV 187
Query: 1029 IFALKIWRHYLYGETCEIYTDHKSLKYIFEQRDLNLRQRRWMELLKDYDCTILYHPGKAN 1088
+FALKIWR YLYG +I+TDHKSLKYIF Q +LNLRQRRWME + DYD I Y+PGKAN
Sbjct: 188 VFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKAN 367
Query: 1089 VVADALSRKSMGSLAHLTAIKRPIVKEFQEIVES-GVQFEIDHSRTLLAHMKFRSTLIDD 1147
+VADALSR+ + A A + +V + + + +L ++ L
Sbjct: 368 LVADALSRRRVDVSAEREA------DDLDGMVRALRLNVLTKATESLGLEAVNQADLFTR 529
Query: 1148 IKQAQSQDSELMKMVDNVRNGKVSNFSVDSEGVLWLKSRICVPNVDDLRRKILEEAHHSS 1207
I+ AQ QD L K+ N R + + +G + + RI VPN L+ +I+ EAH S
Sbjct: 530 IRLAQGQDENLQKVAQNDR----TEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSR 697
Query: 1208 YTIHPGS 1214
+++HPG+
Sbjct: 698 FSVHPGA 718
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 196 bits (497), Expect = 8e-50
Identities = 148/470 (31%), Positives = 224/470 (47%), Gaps = 28/470 (5%)
Frame = +2
Query: 1169 KVSNFSVDSEGVLWLKSRICVPNVDD-------LRRKILEEAHHSSYTIHPGSNKMYQDL 1221
++S +DS L + RI VP DD LR K+++E+H S+ HPG N + +
Sbjct: 68 QISECQLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIV 247
Query: 1222 REFYWWEGMKRDVANFVSKCLVCQQVKAEHQKPAGLLQPIEIPKWKWEGIAMDFVTGLPR 1281
++W G + V FV C VC + Q G L+P+ +P ++MDF+T LP
Sbjct: 248 SRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPP 427
Query: 1282 TQ-KGFDSVWVIIDRLTKSAHFLPVKTTYTASQYAKIYLEEIVSLHGVPISIISDRGAQF 1340
T+ +G +WVI+DRL+KS L T A A+ +L HG+P SI+SDRG+ +
Sbjct: 428 TRGRGSQYLWVIVDRLSKSVT-LEEMDTMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNW 604
Query: 1341 TAQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRACVLDLGGSWDRYLPMME 1400
+FW+ F G LST++HPQTDG +ER Q ++ +LRA V +W LP ++
Sbjct: 605 VGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQ 784
Query: 1401 FAYNNSYQSSIQMAPFEALYGRRCRSPIGWFE-----VGEAKLVGPELIQDAIDKVKLIR 1455
A N + SSI PF +G PI E V E + L++ D I+
Sbjct: 785 LALRNRHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQ 961
Query: 1456 DRLVTAQSRQKSYSDKRRRPLE-FTVGEHVFLRVSPMKGVLRFGKKGKLTPRFIGPFEIL 1514
+V AQ R ++ ++KRR P + + VG+ V+L VS K + K +E+
Sbjct: 962 AEIVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYKS----PRPSKKLDWLHHKYEVT 1129
Query: 1515 ERVGPVAYRLALPPDLSRVHPVFHISMLRKYIYDP---SHVISHK-----------DVQL 1560
V P L +P V+P FH+ +LR+ DP V+ + + ++
Sbjct: 1130 RFVTPHVVELNVP---GTVYPKFHVDLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEV 1300
Query: 1561 DENLSYTEHPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEPEETIR 1610
+E L+ H V +G RR V WKG + TWE + IR
Sbjct: 1301 EEILAARWHQVG---RGRRR-------QALVKWKG--FVDATWEAADAIR 1414
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 89.4 bits (220), Expect(2) = 1e-32
Identities = 45/74 (60%), Positives = 53/74 (70%)
Frame = -1
Query: 712 KGFIRASTSPWGAPVLFVKKKDGSMRLCVDYRQLNKVTIKNKYPLPRIDELFDQLQGAQC 771
K F + S SP GA +LFV+KKDG R+C+DYRQ NKVT KNKYPLPRID LFD++Q
Sbjct: 274 KRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCY 95
Query: 772 FSKIDLRSGYHQLK 785
F IDLR G L+
Sbjct: 94 F*NIDLRLGIINLE 53
Score = 70.9 bits (172), Expect(2) = 1e-32
Identities = 44/100 (44%), Positives = 61/100 (61%)
Frame = -2
Query: 615 NNLISSLRANKLLSRGCQGYLALVRDVQAGEEKLENVPIVCEFSDVFPEELPGIPPDREI 674
N +IS L KL+++GC + V D+++ E V ++ FS VFP+ P IP +REI
Sbjct: 564 NLIISCLNDCKLITKGCLYHKERVMDLESVIPLFEVVLVLKGFS*VFPDNFPVIPLEREI 385
Query: 675 EFSIDLIPGTQPISIPPYRMAPAELKELREQLQDLLDKGF 714
F IDL+ TQ IS PP M ELK+L+ L+D L+KGF
Sbjct: 384 FFCIDLLLDTQLISNPP*LMDRTELKKLKI*LKDSLEKGF 265
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 130 bits (327), Expect = 4e-30
Identities = 66/125 (52%), Positives = 83/125 (65%)
Frame = +2
Query: 1493 GVLRFGKKGKLTPRFIGPFEILERVGPVAYRLALPPDLSRVHPVFHISMLRKYIYDPSHV 1552
GV R K KLT RFIGP++I ERVG VAYR+ LPP L +H VFH+S LRKY+ DPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 1553 ISHKDVQLDENLSYTEHPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEPEETIRGK 1612
I DVQ+ +NL+ PV + D+ V+ LR K+I V+V+W +GE TWE E +
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 1613 YPHLF 1617
YP LF
Sbjct: 362 YPELF 376
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 94.0 bits (232), Expect(2) = 2e-29
Identities = 50/133 (37%), Positives = 79/133 (58%)
Frame = +2
Query: 789 EDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSKS 848
+D+ KTAF T G Y + VM FGL NA + + L+NR+F L + V+IDD+L+ S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 849 REEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSKVEAVQNWP 908
+H HL+ +TL E + +KC F + S FLG+IV++ GI V+P ++ A+ + P
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 909 RPTSVKEIRSFLG 921
P + +E++ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 55.1 bits (131), Expect(2) = 2e-29
Identities = 32/106 (30%), Positives = 52/106 (48%), Gaps = 4/106 (3%)
Frame = +3
Query: 928 RFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLKECLISAPILALPTSGGGYVVYCD 987
RF+ + P +L F W + CEE+F++LK+ L + P+L+ P +G +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 988 ASRVGLGCVLMQHG----KVIAYASRQLKRHEQNYPTHDLEMAAVI 1029
S + VL++ K I Y S+++ E YPT + AVI
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVI 746
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 89.7 bits (221), Expect = 8e-18
Identities = 42/73 (57%), Positives = 57/73 (77%), Gaps = 1/73 (1%)
Frame = +2
Query: 1482 EHVFLRVSPM-KGVLRFGKKGKLTPRFIGPFEILERVGPVAYRLALPPDLSRVHPVFHIS 1540
E V L+V P +G RFGK+GKL+ R+IGPFE+++R+G VAY LALPP LS VHPVFH+S
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1541 MLRKYIYDPSHVI 1553
M ++Y D +++I
Sbjct: 182 MFKRYHGDGNYII 220
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 65.9 bits (159), Expect(3) = 1e-17
Identities = 34/81 (41%), Positives = 44/81 (53%)
Frame = +2
Query: 1342 AQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRACVLDLGGSWDRYLPMMEF 1401
+ FWK GT L +S+A+HP +DGQSE + E LR + W + P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1402 AYNNSYQSSIQMAPFEALYGR 1422
YN SY S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 35.4 bits (80), Expect(3) = 1e-17
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = +1
Query: 1458 LVTAQSRQKSYSDKRRRPLEFTVGEHVFLRVSP 1490
L AQ K +DK+RR EF +GEHV +++ P
Sbjct: 379 LYKAQQTMKHQADKKRRHFEFQLGEHVLVKLQP 477
Score = 27.7 bits (60), Expect(3) = 1e-17
Identities = 11/25 (44%), Positives = 19/25 (76%)
Frame = +2
Query: 1521 AYRLALPPDLSRVHPVFHISMLRKY 1545
AY+L+LP ++V P+FH+S L+ +
Sbjct: 557 AYKLSLP-STAKVPPIFHVSQLKPF 628
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 73.6 bits (179), Expect = 6e-13
Identities = 40/138 (28%), Positives = 72/138 (51%), Gaps = 1/138 (0%)
Frame = +2
Query: 127 LAEFLKLKPPTFSGSSVSEDPQRFIDGLERLWRALGCSDVRAVELTSYQLEGVAHDWFDT 186
L FL+ PPTF G + +++ +ER++R + C + + V+ ++ L A DW+ +
Sbjct: 107 LETFLRNHPPTFKGRYAPDGA*KWLKEIERIFRVMQCFETQKVQFGTHMLAEEADDWWIS 286
Query: 187 VTHGRQMGSSPMAWKEFSKLFMARFLPQSVRDQLAHEFEIFEQTEG-MSVSEYSARFTQL 245
+ + + + W F K F+ R+ P+ VR + E E E +G MSV+EY+A+F +L
Sbjct: 287 LLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGK--KEIEFLELKQGDMSVTEYAAKFVEL 460
Query: 246 SRYAAYCVTEEMRIKKFI 263
+ + + E K I
Sbjct: 461 ATFYPHYSAETAEFSKCI 514
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 63.5 bits (153), Expect = 6e-10
Identities = 45/177 (25%), Positives = 84/177 (47%), Gaps = 4/177 (2%)
Frame = +2
Query: 1199 ILEEAHHSSYTIHPGSNKMYQDLREF-YWWEGMKRDVANFVSKCLVCQQ---VKAEHQKP 1254
IL H S+Y H +K +++ +WW M +D +F+SKC CQ+ + ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 1255 AGLLQPIEIPKWKWEGIAMDFVTGLPRTQKGFDSVWVIIDRLTKSAHFLPVKTTYTASQY 1314
+ +E+ ++ +DF+ P + + V +D ++K + T A+
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFPSSYNN-KYILVAVDYVSKWVEAI-ASPTNDATVV 445
Query: 1315 AKIYLEEIVSLHGVPISIISDRGAQFTAQFWKSFQAALGTRLNLSTAFHPQTDGQSE 1371
K++ I GVP +ISD G+ F + ++ G R ++TA+HPQ +S+
Sbjct: 446 VKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 53.1 bits (126), Expect = 8e-07
Identities = 23/43 (53%), Positives = 34/43 (78%)
Frame = -3
Query: 1284 KGFDSVWVIIDRLTKSAHFLPVKTTYTASQYAKIYLEEIVSLH 1326
K ++S+ V++DRLTKS F+P KT+Y+A YA+I L+EIV +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>CB893783 weakly similar to GP|22830935|dbj hypothetical protein~similar to
gag-pol polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 853
Score = 47.8 bits (112), Expect = 4e-05
Identities = 38/132 (28%), Positives = 65/132 (48%), Gaps = 13/132 (9%)
Frame = +2
Query: 565 VLNMVDFDVILGMDWLSLHHATVDCHNKV---------VKFKPPGEATFSFQGERSWVPN 615
V++M ++LG W HA D H +K P F +G++ + P
Sbjct: 461 VISMDACHMLLGRPWQYDRHALYDGHANTYTFVKYGVKIKLVPLPPNAFD-EGKKDFKPI 637
Query: 616 NLISSLRANKLLSRGCQGYLALVRDVQAGEEKL---ENVPIVCEFSDVFPEELP-GIPPD 671
+ S K+ ++ Q ++L+ V++ EE E ++ +F+DV P E+P G+PP
Sbjct: 638 VSLVSKEPFKVTTKDIQD-MSLILLVKSNEESTIQKEVEHLLVDFTDVVPSEIPSGLPPM 814
Query: 672 REIEFSIDLIPG 683
R+I+ +ID IPG
Sbjct: 815 RDIQHAIDFIPG 850
>BG644717
Length = 267
Score = 45.8 bits (107), Expect = 1e-04
Identities = 21/46 (45%), Positives = 30/46 (64%)
Frame = -2
Query: 136 PTFSGSSVSEDPQRFIDGLERLWRALGCSDVRAVELTSYQLEGVAH 181
P F GS ++EDPQ F+D +++++ + S VEL SYQL VAH
Sbjct: 260 PEFLGSQINEDPQNFLDEIKKIFEVMHVSGNDLVELASYQL*DVAH 123
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 43.1 bits (100), Expect = 9e-04
Identities = 21/67 (31%), Positives = 39/67 (57%)
Frame = -2
Query: 1326 HGVPISIISDRGAQFTAQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRACV 1385
HG+P I++D G+ F + ++ F RLN ++ +PQ++GQ+E + +I+ D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 1386 LDLGGSW 1392
G W
Sbjct: 505 DLKKGCW 485
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 43.1 bits (100), Expect = 9e-04
Identities = 25/60 (41%), Positives = 34/60 (56%), Gaps = 5/60 (8%)
Frame = +1
Query: 992 GLGCVLMQ-----HGKVIAYASRQLKRHEQNYPTHDLEMAAVIFALKIWRHYLYGETCEI 1046
G+G L Q H IAYASR L E+NY + E A I+A++ +RHYL+G E+
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 42.7 bits (99), Expect = 0.001
Identities = 19/54 (35%), Positives = 28/54 (51%)
Frame = -1
Query: 1564 LSYTEHPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEPEETIRGKYPHLF 1617
L PV ++D+ + +R K I VK++W EE TWE E ++ YP F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Frame = -2
Query: 1027 AVIFALKIWRHYLYGETCEIYTDHKSLKYIFEQRDLNLRQRRWMELLKDYDCTILYHPGK 1086
A+ +A K RHY+ T + + +KYIFE+ L R RW LL +YD I Y K
Sbjct: 623 ALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYD--IEYRSQK 450
Query: 1087 A---NVVADALSRKSM 1099
A +++AD L+ + +
Sbjct: 449 AIKGSILADHLAHQPL 402
>TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protein {Glycine
max}, partial (35%)
Length = 1552
Score = 39.3 bits (90), Expect = 0.013
Identities = 23/51 (45%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Frame = +2
Query: 1 MPPRNRNRPRREVVDESNAP--PQGTRHRGRGRARGGRGVNVRGRGVNAGR 49
+ PR+R RPR+ V + P RGRGR RG GV RGRG GR
Sbjct: 980 LTPRSRRRPRKNVNAVAAVPVAAVAATGRGRGRGRGRGGVAGRGRGGGRGR 1132
>BG644743
Length = 122
Score = 39.3 bits (90), Expect = 0.013
Identities = 15/33 (45%), Positives = 25/33 (75%)
Frame = +1
Query: 1484 VFLRVSPMKGVLRFGKKGKLTPRFIGPFEILER 1516
V+ +SPM GV+RFGKK K +P ++G +++L +
Sbjct: 16 VY*TISPMNGVMRFGKKEKFSPCYVGLYDVLNQ 114
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,772,121
Number of Sequences: 36976
Number of extensions: 686483
Number of successful extensions: 3654
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 3453
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3617
length of query: 1621
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1512
effective length of database: 4,984,343
effective search space: 7536326616
effective search space used: 7536326616
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC126790.10


