

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.13 - phase: 0
(916 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
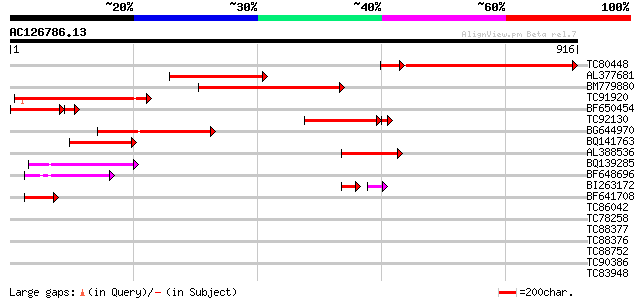
Score E
Sequences producing significant alignments: (bits) Value
TC80448 similar to PIR|C96495|C96495 probable ligand-gated ion c... 564 e-179
AL377681 weakly similar to PIR|T51131|T511 ligand gated channel-... 324 1e-88
BM779880 weakly similar to GP|13160471|gb putative glutamate rec... 307 1e-83
TC91920 weakly similar to PIR|T45779|T45779 probable glutamate r... 239 5e-63
BF650454 similar to PIR|C96495|C964 probable ligand-gated ion ch... 179 9e-52
TC92130 similar to PIR|C96495|C96495 probable ligand-gated ion c... 182 3e-51
BG644970 weakly similar to GP|20197431|gb| putative ligand-gated... 172 7e-43
BQ141763 weakly similar to PIR|T45779|T4 probable glutamate rece... 149 6e-36
AL388536 weakly similar to PIR|C96495|C96 probable ligand-gated ... 132 6e-31
BQ139285 weakly similar to PIR|T05099|T05 hypothetical protein F... 83 5e-16
BF648696 54 3e-07
BI263172 weakly similar to PIR|T45779|T45 probable glutamate rec... 34 2e-05
BF641708 weakly similar to PIR|T51137|T511 ionotropic glutamate ... 47 3e-05
TC86042 similar to GP|20466712|gb|AAM20673.1 putative SF16 prote... 34 0.22
TC78258 similar to GP|21593869|gb|AAM65836.1 polygalacturonase i... 33 0.65
TC88377 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidop... 32 1.1
TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidop... 31 2.5
TC88752 Narf-like protein [Medicago truncatula] 30 5.5
TC90386 weakly similar to GP|5882724|gb|AAD55277.1| F25A4.4 {Ara... 29 7.2
TC83948 29 9.4
>TC80448 similar to PIR|C96495|C96495 probable ligand-gated ion channel
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1200
Score = 564 bits (1453), Expect(2) = e-179
Identities = 278/287 (96%), Positives = 283/287 (97%)
Frame = +3
Query: 630 LSTLFFSHICRREHNEYSWPRCSTHMVVCGFDN*FQLHC*FNIHTHGAAVIFTYQWH*KL 689
++++FF +REHNEYSWPRCSTHMVVCGFDN*FQLHC*FNIHTHGAAVIFTYQWH*KL
Sbjct: 93 VNSIFFP---QREHNEYSWPRCSTHMVVCGFDN*FQLHC*FNIHTHGAAVIFTYQWH*KL 263
Query: 690 KS***TNRVSSGFIC*TLLD*RYWNIQI*TCSSWISRRICQGTPAWS**RRCCCYC**TA 749
KS***TNRVSSGFIC*TLLD*RYWNIQI*TCSSWISRRICQGTPAWS**RRCCCYC**TA
Sbjct: 264 KS***TNRVSSGFIC*TLLD*RYWNIQI*TCSSWISRRICQGTPAWS**RRCCCYC**TA 443
Query: 750 LCRNILINPMHIQDCRSGVHQKWLGVCIPSRLPFGGGFVNCHPTTF*NWGSATDS**VDD 809
LCRNILINPMHIQDCRSGVHQKWLGVCIPSRLPFGGGFVNCHPTTF*NWGSATDS**VDD
Sbjct: 444 LCRNILINPMHIQDCRSGVHQKWLGVCIPSRLPFGGGFVNCHPTTF*NWGSATDS**VDD 623
Query: 810 AKHL*FR*Y*N*IRSSSAQKFLGSLHHLRGGLLYCSRHIFFADYVTGTTFHSSRISF*CW 869
AKHL*FR*Y*N*IRSSSAQKFLGSLHHLRGGLLYCSRHIFFADYVTGTTFHSSRISF*CW
Sbjct: 624 AKHL*FR*Y*N*IRSSSAQKFLGSLHHLRGGLLYCSRHIFFADYVTGTTFHSSRISF*CW 803
Query: 870 PTSKISFTHRREERSIEE*KKKEEWR*NIT*RSTGKATEEDTKSNGS 916
PTSKISFTHRREERSIEE*KKKEEWR*NIT*RSTGKATEEDTKSNGS
Sbjct: 804 PTSKISFTHRREERSIEE*KKKEEWR*NIT*RSTGKATEEDTKSNGS 944
Score = 83.2 bits (204), Expect(2) = e-179
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = +1
Query: 600 GIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
GIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH
Sbjct: 1 GIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 114
>AL377681 weakly similar to PIR|T51131|T511 ligand gated channel-like protein
[imported] - rape, partial (6%)
Length = 475
Score = 324 bits (830), Expect = 1e-88
Identities = 158/158 (100%), Positives = 158/158 (100%)
Frame = +1
Query: 259 WLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAY 318
WLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAY
Sbjct: 1 WLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAY 180
Query: 319 DTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFV 378
DTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFV
Sbjct: 181 DTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFV 360
Query: 379 GLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNY 416
GLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNY
Sbjct: 361 GLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNY 474
>BM779880 weakly similar to GP|13160471|gb putative glutamate receptor
like-protein {Arabidopsis thaliana}, partial (11%)
Length = 721
Score = 307 bits (787), Expect = 1e-83
Identities = 142/236 (60%), Positives = 178/236 (75%)
Frame = +1
Query: 306 GSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNG 365
G LGL++YG+ AYDT++++A+A+D F QG ++ ++ L+ + L+LDA+ IF+ G
Sbjct: 13 GPLGLSTYGIFAYDTIYVLARALDTFLKQGNQITFSSDPKLNQPRGDSLHLDAVKIFNEG 192
Query: 366 TLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPE 425
LL +I N G++GP + + +L PAY+IINV+G G RRVGYWSNYSGLS++ PE
Sbjct: 193 NLLRKSIYEVNMTGVTGPFRYTPDGNLANPAYEIINVIGTGTRRVGYWSNYSGLSVIPPE 372
Query: 426 TLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKG 485
TLY+ PPNRS NQ L TV WPGETT RPRGWVFPNNGK L+IGVP R SYREFVS V+
Sbjct: 373 TLYSKPPNRSIDNQKLLTVFWPGETTQRPRGWVFPNNGKLLKIGVPRRTSYREFVSQVQS 552
Query: 486 TDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVG 541
TD FKGFC+DVF++AVNLLPYAVPY+FVP+GDG NPS TE V IT G FD AVG
Sbjct: 553 TDTFKGFCIDVFLSAVNLLPYAVPYKFVPYGDGQNNPSNTELVRLITAGVFDAAVG 720
>TC91920 weakly similar to PIR|T45779|T45779 probable glutamate receptor -
Arabidopsis thaliana, partial (14%)
Length = 868
Score = 239 bits (609), Expect = 5e-63
Identities = 118/229 (51%), Positives = 172/229 (74%), Gaps = 7/229 (3%)
Frame = +3
Query: 8 LWLVFLFMLPYL------EQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSN 61
+W V L +L L + +++ P FVNIG +++F++S+G++ K+A E AV D+NS+
Sbjct: 135 VWFVVLMVLYNLMFSSTVAGLDNSTVPPFVNIGVLYSFNTSVGRMVKIAAEAAVADINSD 314
Query: 62 SSILHSTQLVLHMQT-SNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRV 120
+IL +T+L L +Q S GF + +AL+ M T +AI+GPQ+S +H+++H+ANEL+V
Sbjct: 315 PTILGNTKLNLSLQEDSKYRGFLSIAEALQLMATQTVAIIGPQTSTTAHVISHIANELQV 494
Query: 121 PMLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGV 180
P+LSF ATDPTLSSLQFPFF+RT+ +D++QMTA+A+I+ YGW+EVIT+Y DDD+GRNG+
Sbjct: 495 PLLSFTATDPTLSSLQFPFFLRTSFNDIFQMTAIADIVSHYGWREVITVYGDDDHGRNGI 674
Query: 181 SALDDALAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVH 229
SAL D LAERRC+IS+K + PD EIT++LV VA+ +SRIIV+H
Sbjct: 675 SALGDKLAERRCKISFKAAMT--PDATSEEITDVLVQVALAESRIIVLH 815
>BF650454 similar to PIR|C96495|C964 probable ligand-gated ion channel
[imported] - Arabidopsis thaliana, partial (3%)
Length = 664
Score = 179 bits (455), Expect(2) = 9e-52
Identities = 87/88 (98%), Positives = 87/88 (98%)
Frame = +2
Query: 1 MNFYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNS 60
MNFYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNS
Sbjct: 323 MNFYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNS 502
Query: 61 NSSILHSTQLVLHMQTSNCSGFDGMIQA 88
NSSILHSTQLVLHMQT NCSGFDGMIQA
Sbjct: 503 NSSILHSTQLVLHMQTXNCSGFDGMIQA 586
Score = 43.5 bits (101), Expect(2) = 9e-52
Identities = 21/25 (84%), Positives = 22/25 (88%)
Frame = +3
Query: 89 LRFMETDVIAILGPQSSVVSHIVAH 113
LR METDV+ ILGPQSSVVSHI AH
Sbjct: 588 LRVMETDVLPILGPQSSVVSHIRAH 662
>TC92130 similar to PIR|C96495|C96495 probable ligand-gated ion channel
[imported] - Arabidopsis thaliana, partial (10%)
Length = 878
Score = 182 bits (461), Expect(2) = 3e-51
Identities = 83/125 (66%), Positives = 100/125 (79%)
Frame = +3
Query: 476 YREFVSPVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGY 535
YR+FVS V GTD F+GFC+DVF++A+NLLPYAVPY+F+P+GDG NPS TE V +ITTG
Sbjct: 3 YRQFVSQVPGTDTFQGFCIDVFLSAINLLPYAVPYKFIPYGDGKNNPSNTELVRRITTGE 182
Query: 536 FDGAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACF 595
+DGAVGDIAI T RT++VDFTQPY SGLVVVAP ++ + +FL PFTP MW VTA F
Sbjct: 183 YDGAVGDIAITTTRTKMVDFTQPYIESGLVVVAPVRETETSALAFLAPFTPRMWFVTALF 362
Query: 596 FFFVG 600
F VG
Sbjct: 363 FIIVG 377
Score = 39.7 bits (91), Expect(2) = 3e-51
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = +1
Query: 601 IVVWILEHRVNDEFRGSP 618
+VVWILEHRVNDEFRG P
Sbjct: 379 LVVWILEHRVNDEFRGPP 432
>BG644970 weakly similar to GP|20197431|gb| putative ligand-gated ion channel
subunit {Arabidopsis thaliana}, partial (7%)
Length = 810
Score = 172 bits (435), Expect = 7e-43
Identities = 90/194 (46%), Positives = 122/194 (62%), Gaps = 3/194 (1%)
Frame = +2
Query: 142 RTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIK 201
RT SD QM A+A +IDF GWKEVI I++DDDYGRNG+SAL D L +RR ++++K+ +
Sbjct: 5 RTIQSDSEQMAAMANLIDFNGWKEVIVIFLDDDYGRNGISALSDELEKRRLKLAHKLPLS 184
Query: 202 SGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLS 261
D+D EIT LL + R+ VVH + + IF +A L MM YVW+ATDWLS
Sbjct: 185 IHYDLD--EITKLLNQSRVYSPRVFVVHVNPDPRLRIFSIARKLQMMTSDYVWLATDWLS 358
Query: 262 TVLDS-TSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGG--SLGLNSYGLHAY 318
S +S ++ ++G + LRQH PD+ +K+ F S+W + G + LNSYG AY
Sbjct: 359 ATSHSFSSANQNSLSIVEGVVALRQHVPDSRKKRDFISRWKKMQKGVANTSLNSYGFFAY 538
Query: 319 DTVWLVAQAIDNFF 332
DTVW VA +ID F
Sbjct: 539 DTVWTVAHSIDK*F 580
>BQ141763 weakly similar to PIR|T45779|T4 probable glutamate receptor -
Arabidopsis thaliana, partial (10%)
Length = 797
Score = 149 bits (375), Expect = 6e-36
Identities = 65/108 (60%), Positives = 87/108 (80%)
Frame = +2
Query: 97 IAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAE 156
+AI+GPQ+S +H+++H+ NEL+VP+LSF ATDPTLS LQFPFF+RT +D++QMTA+A+
Sbjct: 44 VAIIGPQTSTTAHVISHIPNELQVPLLSFTATDPTLSLLQFPFFLRTFFNDIFQMTAIAD 223
Query: 157 IIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGP 204
I YGW+EVIT+Y DDD+GRNG+S L D LA+RRC IS K + P
Sbjct: 224 IASHYGWREVITVYGDDDHGRNGISLLGDKLADRRCTISPKAALTPDP 367
>AL388536 weakly similar to PIR|C96495|C96 probable ligand-gated ion channel
[imported] - Arabidopsis thaliana, partial (14%)
Length = 486
Score = 132 bits (332), Expect = 6e-31
Identities = 59/98 (60%), Positives = 73/98 (74%)
Frame = +2
Query: 537 DGAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFF 596
+ GDIAIVTNRT+I DF+QP+A+S LVVVAP S W FL+PF+P MW + F
Sbjct: 23 EAVAGDIAIVTNRTKIADFSQPFASSSLVVVAPINSSKSNAWVFLKPFSPDMWCIIVASF 202
Query: 597 FFVGIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLF 634
+G+V+WILEHRVND+FRG PK+Q VT+ FSLSTLF
Sbjct: 203 MMIGVVIWILEHRVNDDFRGPPKRQLVTMFMFSLSTLF 316
>BQ139285 weakly similar to PIR|T05099|T05 hypothetical protein F28M20.100 -
Arabidopsis thaliana, partial (17%)
Length = 682
Score = 82.8 bits (203), Expect = 5e-16
Identities = 48/180 (26%), Positives = 98/180 (53%), Gaps = 3/180 (1%)
Frame = +1
Query: 31 VNIGAIFTFDSS--IGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDGMIQA 88
V +GA+ S+ +GK+ + ++ D + S + T++ L ++ S+ A
Sbjct: 133 VKVGAVIDISSNETVGKIGLSCINMSLSDFYLSHSH-YKTRIQLIVRDSHRDVVAAAAHA 309
Query: 89 LRFMETDVI-AILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLSD 147
L ++ + + AI+GP +++ ++ V + ++ VP+++F+AT P+L+SLQ +F + + +D
Sbjct: 310 LDLIKNEEVHAIMGPITTMEANFVIQLGDKAHVPIVTFSATSPSLASLQSSYFFQISQND 489
Query: 148 LYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDVD 207
Q+ A+ II +GWK+V+ IYVD+ +G + L L + ++ Y I + D
Sbjct: 490 STQVKAITSIIQAFGWKQVVPIYVDNSFGEXLIPYLTSVLQQAYIQVPYLSAISLSANDD 669
>BF648696
Length = 653
Score = 53.9 bits (128), Expect = 3e-07
Identities = 42/148 (28%), Positives = 72/148 (48%), Gaps = 3/148 (2%)
Frame = +2
Query: 25 NSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDG 84
N ++IG I +S IGK +LA + SN+S + +LVL+ Q S
Sbjct: 35 NEYSKVISIGVIVDVNSRIGKEQELA--EIASQSYSNTS--KNYKLVLYFQNSTKDTLKA 202
Query: 85 MIQALRFMETDVI-AILGPQSSVVSHIVAHVANELRVPMLSFAATD--PTLSSLQFPFFV 141
+ A + + I+G + + I+ + ++ +VP++SFAA P L + ++PF V
Sbjct: 203 IKIAEEMINVQKVQVIIGMHTWPEAAIMEDIGSKAQVPIISFAAPTITPPLMNNRWPFLV 382
Query: 142 RTTLSDLYQMTAVAEIIDFYGWKEVITI 169
R + + +AEI+ Y WK V+ I
Sbjct: 383 RLANNGTTYIKCIAEIVHAYCWKRVVVI 466
>BI263172 weakly similar to PIR|T45779|T45 probable glutamate receptor -
Arabidopsis thaliana, partial (2%)
Length = 683
Score = 33.9 bits (76), Expect(2) = 2e-05
Identities = 14/32 (43%), Positives = 20/32 (61%)
Frame = +1
Query: 536 FDGAVGDIAIVTNRTRIVDFTQPYAASGLVVV 567
+D D+ I+ R+R V FTQPY SGL ++
Sbjct: 424 YDAIGADVTILAERSRNVSFTQPYTESGLSLI 519
Score = 33.1 bits (74), Expect(2) = 2e-05
Identities = 11/33 (33%), Positives = 18/33 (54%)
Frame = +2
Query: 578 WSFLQPFTPFMWIVTACFFFFVGIVVWILEHRV 610
W ++PF +WI T + I++W LEH +
Sbjct: 548 WLIMKPFXWEIWIATXGILIYTMIIIWXLEHHL 646
>BF641708 weakly similar to PIR|T51137|T511 ionotropic glutamate receptor
homolog GLR4 [imported] - Arabidopsis thaliana, partial
(4%)
Length = 362
Score = 47.4 bits (111), Expect = 3e-05
Identities = 22/55 (40%), Positives = 36/55 (65%)
Frame = +1
Query: 25 NSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNC 79
+S P+ V IGA+FT DS IG+ A+ ++ A+ DVN+N +IL ++ + +NC
Sbjct: 193 SSXPTVVKIGALFTVDSVIGRSAQQGIKTAIDDVNANXTILPGIKMDVIFHDTNC 357
>TC86042 similar to GP|20466712|gb|AAM20673.1 putative SF16 protein
{Arabidopsis thaliana}, partial (39%)
Length = 2305
Score = 34.3 bits (77), Expect = 0.22
Identities = 30/130 (23%), Positives = 55/130 (42%), Gaps = 18/130 (13%)
Frame = -3
Query: 492 FCVDVFVAAVNLLPY---AVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTN 548
+CV + A N LP+ +V + F+P + + F+ K+ + + V +
Sbjct: 461 WCVKTLLIAANQLPFFPISVAFLFLPERSTERR-QHLYFIFKLVSCHRASKVAKSTVKVT 285
Query: 549 RT----------RIVDFTQPYAASGLVVVAPFKKINSGGWSFL-----QPFTPFMWIVTA 593
+T + F P + L+++ F + S + L Q FT F+W +
Sbjct: 284 KTDQKRRNSLSLSLASFFFPAPHN*LIILLSFSLLESNSFGLLLFLF*QTFT-FLWSL*L 108
Query: 594 CFFFFVGIVV 603
FF FVG++V
Sbjct: 107 IFFVFVGLIV 78
>TC78258 similar to GP|21593869|gb|AAM65836.1 polygalacturonase inhibiting
protein 1 {Arabidopsis thaliana}, partial (18%)
Length = 885
Score = 32.7 bits (73), Expect = 0.65
Identities = 28/103 (27%), Positives = 41/103 (39%)
Frame = -1
Query: 382 GPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHL 441
GP++L RS F ++ +G G +G +N L + P R +
Sbjct: 486 GPLRLLEPRSSFCSLLSLLKELGIGPVTLGISANERKLRYGRSPNSAGSVPERPQPRLMV 307
Query: 442 HTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVK 484
TVI PG RG P + L+ VP+ S R + P K
Sbjct: 306 TTVIRPG------RGLPHPTKLQSLQQSVPLSQSERRPLGPPK 196
>TC88377 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidopsis
thaliana}, partial (23%)
Length = 644
Score = 32.0 bits (71), Expect = 1.1
Identities = 19/60 (31%), Positives = 29/60 (47%)
Frame = +1
Query: 102 PQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFY 161
PQ +V +L +P L T+PTL SL FP F +L +L ++ + I F+
Sbjct: 256 PQGNVPPLPTIPSVPKLTMPPLPSIPTNPTLPSLNFPPFPSNSLPNLPSISTIISSIPFF 435
>TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidopsis
thaliana}, partial (25%)
Length = 706
Score = 30.8 bits (68), Expect = 2.5
Identities = 15/45 (33%), Positives = 25/45 (55%)
Frame = +1
Query: 117 ELRVPMLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFY 161
+L +P L T+PTL SL FP F +L ++ ++ + I F+
Sbjct: 319 KLTMPPLPSIPTNPTLPSLNFPPFPSNSLPNIPSISTIISSIPFF 453
>TC88752 Narf-like protein [Medicago truncatula]
Length = 1962
Score = 29.6 bits (65), Expect = 5.5
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +3
Query: 322 WLVAQAIDNFFSQGGVVSCTNY 343
WL+ + NF SQGG C NY
Sbjct: 1731 WLIGVELSNFGSQGGYFVCNNY 1796
>TC90386 weakly similar to GP|5882724|gb|AAD55277.1| F25A4.4 {Arabidopsis
thaliana}, partial (54%)
Length = 674
Score = 29.3 bits (64), Expect = 7.2
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = +1
Query: 604 WILEHRVNDEFRGSPKQQFVTILWFSLSTLFF 635
W+L+H FR SP + WF +S +FF
Sbjct: 70 WLLDHPSIVSFRWSPALSWGATWWFLISAIFF 165
>TC83948
Length = 708
Score = 28.9 bits (63), Expect = 9.4
Identities = 15/47 (31%), Positives = 25/47 (52%)
Frame = +2
Query: 1 MNFYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVA 47
+NFY+ WL F + +L+ S ++ +FTF S IGK++
Sbjct: 509 LNFYFLWLRFTFHVLRGFLDS------DSDLHSWYLFTFGSMIGKIS 631
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.147 0.498
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,183,348
Number of Sequences: 36976
Number of extensions: 556274
Number of successful extensions: 4419
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 2270
Number of HSP's successfully gapped in prelim test: 219
Number of HSP's that attempted gapping in prelim test: 2056
Number of HSP's gapped (non-prelim): 2638
length of query: 916
length of database: 9,014,727
effective HSP length: 105
effective length of query: 811
effective length of database: 5,132,247
effective search space: 4162252317
effective search space used: 4162252317
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC126786.13


