

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.10 - phase: 0 /pseudo
(1308 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
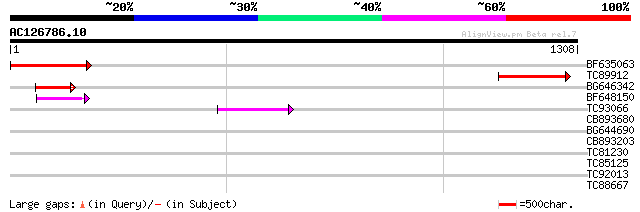
Score E
Sequences producing significant alignments: (bits) Value
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 287 2e-77
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 157 3e-38
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 115 1e-25
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 64 4e-10
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 46 8e-05
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 34 0.32
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 32 1.2
CB893203 32 1.2
TC81230 32 1.6
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 32 2.1
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 31 3.6
TC88667 similar to GP|6728982|gb|AAF26980.1| unknown protein {Ar... 31 3.6
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 287 bits (735), Expect = 2e-77
Identities = 145/186 (77%), Positives = 164/186 (87%)
Frame = -2
Query: 2 MGSK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVS 61
MGSK DIEKFTG NDFGLWKVKM A+LIQQKC +ALKGE + ++ AEKTE+ DKA S
Sbjct: 568 MGSKRDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEVSLPVTMSRAEKTEMVDKARS 389
Query: 62 AIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQ 121
AI++CLGDKVLREV++E TA SMW KL SLYMTKSLAHRQ LKQQLY +RMVESK IMEQ
Sbjct: 388 AIVLCLGDKVLREVAKERTAASMWAKL*SLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQ 209
Query: 122 LTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALR 181
LTEFNKI+D+L NI+V LEDE+KA+ LLCALP+SFE+FKDTMLYGKEGT+TLEEVQAALR
Sbjct: 208 LTEFNKILDDLENIEVQLEDEEKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALR 29
Query: 182 TKELTK 187
TKELTK
Sbjct: 28 TKELTK 11
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 157 bits (396), Expect = 3e-38
Identities = 88/164 (53%), Positives = 109/164 (65%)
Frame = +3
Query: 1129 SFEVGSKIFEWILERWSKVYKVTTR*GCFGGICGCGLCR*RRH*KIFVRFCVYIVRYSGN 1188
SFEVG ++FE + E +V+K ++R FGG+C C LC H KI + FCVY + +
Sbjct: 3 SFEVGVEVFE*VFEEQFEVHKSSSRGRRFGGVC*CRLCGQCGHKKISIGFCVYSLWHDY* 182
Query: 1189 LEGKSTISCSPFNNSSRVHSPC*RGQGSHMVERYDWRNGN*SRVCEDTL**PKCHSFGKS 1248
LEGKSTI NNSS VH C RG+ HMVERYDW N SR+CEDTL** KCHS G+S
Sbjct: 183 LEGKSTIRGDIINNSSGVHCLCRRGERCHMVERYDW*VRNYSRICEDTL**SKCHSLGES 362
Query: 1249 SGIS*KDKTH*HSFALRQRHD*DKGDHDRESGIGRQSSRHVHQI 1292
S +S*+D H*HS AL RHD* K D ++GIGR+S V+Q+
Sbjct: 363 SSVS*ED*AH*HSLALY*RHD*IKRDCGGKNGIGRESGGCVYQV 494
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 115 bits (288), Expect = 1e-25
Identities = 63/94 (67%), Positives = 74/94 (78%)
Frame = +2
Query: 59 AVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPI 118
A SAI++CLGDKVLREV++E TA SM KL+ LYMTKSLAHRQ LKQQLY ++MVESK I
Sbjct: 2 ARSAIVLCLGDKVLREVAKEPTATSMCAKLEYLYMTKSLAHRQFLKQQLYSFKMVESKAI 181
Query: 119 MEQLTEFNKIIDNLANIDVNLEDEDKALHLLCAL 152
E L EFNKII +L NI+V+LED AL + C L
Sbjct: 182 TELLVEFNKIIGDLENIEVHLEDAG-ALMVWCCL 280
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 63.9 bits (154), Expect = 4e-10
Identities = 36/122 (29%), Positives = 68/122 (55%)
Frame = +2
Query: 63 IIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQL 122
I+ + D + +A +W+KL++ YM + ++ L Y+MV++K +MEQL
Sbjct: 212 ILNGMSDSLFDIYQSSPSAKDLWDKLETRYMREDATSKKFLVSHFNNYKMVDNKSVMEQL 391
Query: 123 TEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRT 182
E +I++N ++N+++ ++ LP S+++FK TM + KE I+LE++ LR
Sbjct: 392 YEIERILNNYKQHNMNMDETIIVSSIIDKLPPSWKDFKRTMKHKKE-DISLEQLGNHLRL 568
Query: 183 KE 184
E
Sbjct: 569 XE 574
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 46.2 bits (108), Expect = 8e-05
Identities = 54/178 (30%), Positives = 75/178 (41%), Gaps = 2/178 (1%)
Frame = +3
Query: 479 SFGSLGSSIS*DSWGRFIFHVYR**LF*KSMGLHSEE*K*CF*KIQRMGYTCRKSDWN*T 538
SF LG+ S W ++ Y ** + +GL *K* F IQ + +C SD
Sbjct: 108 SF*PLGTFKSYFLWRTPLYDDYH**FSSEGLGLFFAV*K*DFSHIQEVENSC*NSDREEC 287
Query: 539 ESVEN*QWPEVCFRAV**VLQEERYKEA*NRGIHTSTEWSC*KNEQDFVGACEVYAAGSW 598
E N V +**VL + Y N + T+ C N+QD +YA W
Sbjct: 288 EEAHNR*LIRVL***L**VLHKSWYC*TQNHSKESPTKRCCRTNDQDST*ESSMYALKCW 467
Query: 599 IVQE--FLGRGC*YCSIFD*QMSINRDRSQDTYGGLEWETGRLL*LKSFRSFSVCSCQ 654
+++ LGRG YC S Q + L + L * K+F S+C+CQ
Sbjct: 468 VIELT*SLGRGSIYCMSLGQPFSTFST*LQSSRRYLVR*SC*LF*FKNFWMSSICTCQ 641
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 34.3 bits (77), Expect = 0.32
Identities = 18/48 (37%), Positives = 26/48 (53%), Gaps = 1/48 (2%)
Frame = -1
Query: 903 GCEDRISTWRVGRNYLYATT-RRFCRRQYKSMFVEEIFVWVEAKSKAV 949
GCED IS+WR+ Y++A T R R +E VW + +SK +
Sbjct: 474 GCEDCISSWRLS*GYIHAPT*RILIRSGENGGKTKEEHVWTKTRSKTM 331
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 32.3 bits (72), Expect = 1.2
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 2/67 (2%)
Frame = -1
Query: 902 DGCEDRISTWRVGRNYLYATTRRF--CRRQYKSMFVEEIFVWVEAKSKAVVSSV**VPSK 959
+GCE+ I WR R + T CR + +E +W EA SK++V V ++
Sbjct: 275 NGCEECIY*WRSQRGGVCQATSWI*RCRGTKSCVQIE*DTIWSEASSKSMV*KAVKVSAE 96
Query: 960 GWFCEKQ 966
WF ++Q
Sbjct: 95 EWFQKRQ 75
>CB893203
Length = 800
Score = 32.3 bits (72), Expect = 1.2
Identities = 17/55 (30%), Positives = 27/55 (48%)
Frame = -3
Query: 667 FHGLS*RCERLQTVEDGTWRIKIYYKQGCYF**DPHGDEVQRPGYKLGNGDRENS 721
F GLS RC + T+++G W ++++ C+ PGY L D+ NS
Sbjct: 747 FRGLSDRCPIMLTIDEGNWGPRLHHMLKCW---------ADLPGYHLFVKDKWNS 610
>TC81230
Length = 958
Score = 32.0 bits (71), Expect = 1.6
Identities = 26/123 (21%), Positives = 51/123 (41%), Gaps = 12/123 (9%)
Frame = +1
Query: 74 EVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLA 133
+ R A +W+ L Y L+H+ L + L + +P+ E L + I + L
Sbjct: 454 QFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQPVYEFLAQMEVIWNQLT 633
Query: 134 NIDVNLEDED------------KALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALR 181
+ + +L+D + + L AL +E + + L+ + TLE L+
Sbjct: 634 SCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYEPVRASSLH-QNPLPTLENALPCLK 810
Query: 182 TKE 184
++E
Sbjct: 811 SEE 819
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 31.6 bits (70), Expect = 2.1
Identities = 19/49 (38%), Positives = 24/49 (48%)
Frame = +3
Query: 1159 GICGCGLCR*RRH*KIFVRFCVYIVRYSGNLEGKSTISCSPFNNSSRVH 1207
G+C CR* KI+ CV+ R S L + T CS N+ S VH
Sbjct: 78 GLC*FRFCR*S**KKIYYWLCVHACRRSSKLVVQVTNGCSSVNDRSGVH 224
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 30.8 bits (68), Expect = 3.6
Identities = 16/71 (22%), Positives = 32/71 (44%)
Frame = -2
Query: 168 EGTITLEEVQAALRTKELTKFKELKVDDSGEGLNVSRGRSQNRGKGKGKNSRSKSRSKGD 227
E I+ E ++ + + ++ G G RGR + RG+G+G+ + S ++G+
Sbjct: 344 ESEISSSESESGIGGSPAMRGPSMRGRGGGRGRGGGRGRGRGRGRGRGRGRGN*SVAEGE 165
Query: 228 GNKTQYKCFIC 238
+ C C
Sbjct: 164 SLMPRADCESC 132
>TC88667 similar to GP|6728982|gb|AAF26980.1| unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1018
Score = 30.8 bits (68), Expect = 3.6
Identities = 13/28 (46%), Positives = 20/28 (71%)
Frame = -1
Query: 137 VNLEDEDKALHLLCALPRSFENFKDTML 164
+NLE +DK +H + AL RS ++ + TML
Sbjct: 946 LNLEPKDKRMHAIAALKRSLKSLRITML 863
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.360 0.161 0.601
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,865,664
Number of Sequences: 36976
Number of extensions: 600697
Number of successful extensions: 5656
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1612
Number of HSP's successfully gapped in prelim test: 242
Number of HSP's that attempted gapping in prelim test: 3882
Number of HSP's gapped (non-prelim): 2068
length of query: 1308
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1200
effective length of database: 5,021,319
effective search space: 6025582800
effective search space used: 6025582800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC126786.10


