

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
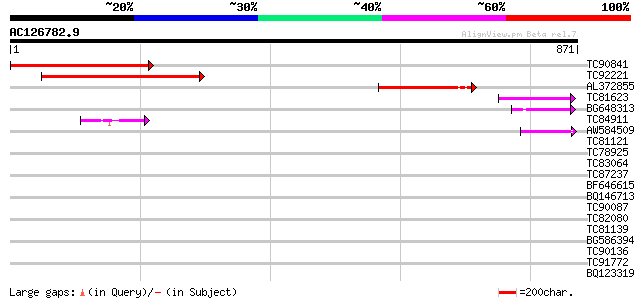
Query= AC126782.9 + phase: 1 /pseudo
(871 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90841 similar to GP|9758283|dbj|BAB08807.1 gene_id:MLE2.5~simi... 432 e-121
TC92221 similar to GP|9758283|dbj|BAB08807.1 gene_id:MLE2.5~simi... 405 e-113
AL372855 similar to GP|9758284|dbj| gene_id:MLE2.6~pir||T01257~s... 221 9e-58
TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcri... 53 4e-07
BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Ara... 50 3e-06
TC84911 similar to PIR|G96635|G96635 hypothetical protein T7P1.1... 44 3e-04
AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pi... 43 5e-04
TC81121 homologue to GP|18182311|gb|AAL65125.1 GT-2 factor {Glyc... 40 0.003
TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Ar... 39 0.011
TC83064 similar to GP|14423452|gb|AAK62408.1 Unknown protein {Ar... 37 0.025
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 35 0.095
BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabido... 35 0.12
BQ146713 similar to GP|9758309|dbj| contains similarity to AAA-t... 34 0.28
TC90087 similar to PIR|T05841|T05841 spliceosome-associated prot... 33 0.36
TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity... 33 0.47
TC81139 similar to PIR|D86427|D86427 hypothetical protein AAG309... 32 0.80
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 32 1.0
TC90136 weakly similar to PIR|T04494|T04494 hypothetical protein... 31 1.8
TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown prot... 31 2.3
BQ123319 similar to GP|10140692|gb| kinesin-like protein {Oryza ... 30 3.0
>TC90841 similar to GP|9758283|dbj|BAB08807.1 gene_id:MLE2.5~similar to
unknown protein~sp|P54122 {Arabidopsis thaliana},
partial (27%)
Length = 658
Score = 432 bits (1110), Expect = e-121
Identities = 218/220 (99%), Positives = 218/220 (99%)
Frame = +1
Query: 1 EQNLHFPLSTI*FHLSIA*HSIVRVFSFLTKMATLTSLPPLPHSLLSLRSKPTRLSVSAS 60
EQNLHFPLSTI*FHLSIA*HSIVRVFSFLTKMATLTSLPPLPHSLLSLRSKPTRLSVSAS
Sbjct: 1 EQNLHFPLSTI*FHLSIA*HSIVRVFSFLTKMATLTSLPPLPHSLLSLRSKPTRLSVSAS 180
Query: 61 ALSASVGNDGSTSRVPQKRRRRIEGPRKSMEDSVQRRMEQFYEGNDGPPLRVLPIGGLGE 120
ALSAS GNDGSTSRVPQKRRRRIEGPRKSMEDSVQRRMEQFYEGNDGPPLRVLPIGGLGE
Sbjct: 181 ALSAS-GNDGSTSRVPQKRRRRIEGPRKSMEDSVQRRMEQFYEGNDGPPLRVLPIGGLGE 357
Query: 121 IGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDH 180
IGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDH
Sbjct: 358 IGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDH 537
Query: 181 IGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPS 220
IGALPWVIPALDSNTPIFASSFTMELIKKRLK HGIFLPS
Sbjct: 538 IGALPWVIPALDSNTPIFASSFTMELIKKRLKXHGIFLPS 657
>TC92221 similar to GP|9758283|dbj|BAB08807.1 gene_id:MLE2.5~similar to
unknown protein~sp|P54122 {Arabidopsis thaliana},
partial (37%)
Length = 956
Score = 405 bits (1040), Expect = e-113
Identities = 196/251 (78%), Positives = 215/251 (85%)
Frame = +3
Query: 49 RSKPTRLSVSASALSASVGNDGSTSRVPQKRRRRIEGPRKSMEDSVQRRMEQFYEGNDGP 108
R PTR S S ++ D ++V KR RRIEGPRKSMEDSVQR+MEQFYEG+DGP
Sbjct: 204 RLHPTRRSHYCRFRSNALPRDTDGAKVVHKRPRRIEGPRKSMEDSVQRKMEQFYEGSDGP 383
Query: 109 PLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKI 168
PLRVLPIGGLGEIGMNCMLVGNHDRYIL+DAG+MFP D+LGVQKIIPDTTFI+KWSHKI
Sbjct: 384 PLRVLPIGGLGEIGMNCMLVGNHDRYILVDAGVMFPGDDELGVQKIIPDTTFIKKWSHKI 563
Query: 169 EALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTK 228
EA+VITHGHEDHIGALPWVIP LDS TP+FASSFTMELI+KRLK+HGIF+PSRLK+FRT+
Sbjct: 564 EAVVITHGHEDHIGALPWVIPMLDSQTPVFASSFTMELIRKRLKDHGIFVPSRLKVFRTR 743
Query: 229 NKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEEL 288
KFVAGPFEIEPI V+HSIPDCCGLVLRCSDGTILHTGDWKIDETPL GKVFD
Sbjct: 744 KKFVAGPFEIEPITVSHSIPDCCGLVLRCSDGTILHTGDWKIDETPLXGKVFDPGSSXGT 923
Query: 289 SKEGVTLMMSD 299
+E V L MSD
Sbjct: 924 VQEXVXLXMSD 956
>AL372855 similar to GP|9758284|dbj| gene_id:MLE2.6~pir||T01257~similar to
unknown protein {Arabidopsis thaliana}, partial (28%)
Length = 439
Score = 221 bits (563), Expect = 9e-58
Identities = 117/150 (78%), Positives = 129/150 (86%)
Frame = +1
Query: 567 DERMRIALDGIIVVSMEIFRPKNLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHA 626
DER+RIALDGIIVVSME+ R ++L+S N LKGKIRITTRCLWLDKGKLLDAL+KAAHA
Sbjct: 1 DERLRIALDGIIVVSMEVCRAQSLDSSVENPLKGKIRITTRCLWLDKGKLLDALHKAAHA 180
Query: 627 ALSSCPVKSPLPHMERTVSEVLRKMVRKYSGKRPEVIAIAIENPGAVFADEINTKLSGKS 686
+LSSCPV PL HME+TVSE+LRKMVRKYSGKRPEVIA+AIENPGAV A EINTKLSGKS
Sbjct: 181 SLSSCPVNCPLAHMEKTVSEMLRKMVRKYSGKRPEVIAVAIENPGAVLATEINTKLSGKS 360
Query: 687 QVGPGISTFRRSVDEHRKENQSTALQIRDD 716
VG GISTFR V KENQST +Q+R D
Sbjct: 361 YVG-GISTFRNVV---HKENQSTKMQMRGD 438
>TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcription
factor [imported] - Arabidopsis thaliana, partial (47%)
Length = 847
Score = 53.1 bits (126), Expect = 4e-07
Identities = 28/118 (23%), Positives = 54/118 (45%)
Frame = +1
Query: 751 FIASSVEKSIKANNGYVSRKEHKSNTKQDDSEDIDEAKSEEMSDSEPESSKSEKKNKWKT 810
F ++S+E ++ ++H+ + +Q + + S +++ +W
Sbjct: 124 FYSTSMEGHHLQHHQQHQHQQHQQHHQQHQQRQHQQHAHNIIGTSSVNVDVTDRFPQWSI 303
Query: 811 EEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSLLADGISRSPGQCKSLWTSLALKYE 868
+E K+ + +RS+L F K LWE IS ++ G RS QCK W +L +Y+
Sbjct: 304 QETKEFLMIRSELDQTFMETKRNKQLWEVISNTMKEKGYHRSAEQCKCKWKNLVTRYK 477
>BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Arabidopsis
thaliana}, partial (53%)
Length = 815
Score = 50.4 bits (119), Expect = 3e-06
Identities = 31/101 (30%), Positives = 48/101 (46%), Gaps = 3/101 (2%)
Frame = +1
Query: 771 EHKSNTKQDDSEDIDEAKSEEMSDSEPESS-KSEKKNK--WKTEEVKKLIDLRSDLRDRF 827
+H + +Q + I A+S S +PE K+ KK W +E + LI LR ++ F
Sbjct: 49 QHHHHLQQQQPQMILTAES---SGDDPEMEIKAPKKRAETWVQDETRSLIGLRREMDSLF 219
Query: 828 KVVKGRMALWEEISQSLLADGISRSPGQCKSLWTSLALKYE 868
K LWE+IS + G RSP C W +L +++
Sbjct: 220 NTSKSNKHLWEQISAKMREKGFDRSPTMCTDKWRNLLKEFK 342
>TC84911 similar to PIR|G96635|G96635 hypothetical protein T7P1.15
[imported] - Arabidopsis thaliana, partial (24%)
Length = 617
Score = 43.5 bits (101), Expect = 3e-04
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 4/109 (3%)
Frame = +3
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGV----QKIIPDTTFIRKWS 165
L V P+G E+G +C+ + + +L D GI P Y + +I P T
Sbjct: 63 LIVTPLGAGNEVGRSCVYMTYKGKTVLFDCGI-HPGYSGMAALPYFDEIDPST------- 218
Query: 166 HKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEH 214
++ L+ITH H DH +LP+ + +F + T + K L ++
Sbjct: 219 --VDVLLITHFHLDHAASLPYFLEKTTFKGRVFMTYATKAIYKLLLSDY 359
>AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pisum
sativum}, partial (22%)
Length = 694
Score = 43.1 bits (100), Expect = 5e-04
Identities = 24/86 (27%), Positives = 43/86 (49%)
Frame = +2
Query: 785 DEAKSEEMSDSEPESSKSEKKNKWKTEEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSL 844
D+ + S +E +S N+W +E L+ +RSD+ FK + LW+E+S+ +
Sbjct: 128 DDERGGGSSRNEEGVDRSFGGNRWPRQETLALLKIRSDMDGAFKDASVKGPLWDEVSRKM 307
Query: 845 LADGISRSPGQCKSLWTSLALKYEVR 870
G R+ +CK + ++ KY R
Sbjct: 308 ADLGYQRNSKKCKEKFENV-YKYHKR 382
>TC81121 homologue to GP|18182311|gb|AAL65125.1 GT-2 factor {Glycine max},
partial (37%)
Length = 794
Score = 40.4 bits (93), Expect = 0.003
Identities = 22/65 (33%), Positives = 35/65 (53%)
Frame = +3
Query: 806 NKWKTEEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSLLADGISRSPGQCKSLWTSLAL 865
N+W +E L+ +RSD+ F+ + LWEE+S+ L G RS +CK + ++
Sbjct: 291 NRWPRQETLALLKIRSDMDGVFRDSSLKGPLWEEVSRKLADLGYHRSSKKCKEKFENV-Y 467
Query: 866 KYEVR 870
KY R
Sbjct: 468 KYHKR 482
>TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 1274
Score = 38.5 bits (88), Expect = 0.011
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Frame = +1
Query: 807 KWKTEEVKKLIDLRSDLRDRFKVVKGRMAL------WEEISQSLLADGISRSPGQCKSLW 860
+W +E+ LI +SD RFK + W +S G++R P QC+ W
Sbjct: 211 RWTRQEILVLIQGKSDAESRFKPGRNGSGFGSSEPKWALVSSYCKKHGVNRGPIQCRKRW 390
Query: 861 TSLALKYE 868
++LA Y+
Sbjct: 391 SNLAGDYK 414
>TC83064 similar to GP|14423452|gb|AAK62408.1 Unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 571
Score = 37.4 bits (85), Expect = 0.025
Identities = 20/69 (28%), Positives = 33/69 (46%), Gaps = 6/69 (8%)
Frame = +1
Query: 801 KSEKKNKWKTEEVKKLIDLRSDL------RDRFKVVKGRMALWEEISQSLLADGISRSPG 854
K E + +W +EV LI+L+S + + + LWE IS+ + G RS
Sbjct: 103 KLEDRRRWPRDEVLALINLKSTTSVINRSNNNVEGNSNKGPLWERISEGMFELGYKRSAK 282
Query: 855 QCKSLWTSL 863
+CK W ++
Sbjct: 283 RCKEKWENI 309
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 35.4 bits (80), Expect = 0.095
Identities = 18/44 (40%), Positives = 30/44 (67%)
Frame = +1
Query: 771 EHKSNTKQDDSEDIDEAKSEEMSDSEPESSKSEKKNKWKTEEVK 814
E KS+ + ++S++ E+ SEE S E E +K E+K+K + EE+K
Sbjct: 55 EEKSHLENEESKETGES-SEEKSHLENEENKDEEKSKQENEEIK 183
>BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabidopsis
thaliana, partial (16%)
Length = 654
Score = 35.0 bits (79), Expect = 0.12
Identities = 17/63 (26%), Positives = 35/63 (54%), Gaps = 1/63 (1%)
Frame = +1
Query: 807 KWKTEEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSLLAD-GISRSPGQCKSLWTSLAL 865
+W +E L+++RS L +FK + LW+++S+ + + G RS +C+ + +L
Sbjct: 346 RWPRQETLTLLEIRSRLDPQFKEATQKGPLWDQLSRIMCEEHGYQRSGKKCREKFENLYK 525
Query: 866 KYE 868
Y+
Sbjct: 526 YYK 534
>BQ146713 similar to GP|9758309|dbj| contains similarity to AAA-type
ATPase~gene_id:MUA2.5 {Arabidopsis thaliana}, partial
(5%)
Length = 571
Score = 33.9 bits (76), Expect = 0.28
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +3
Query: 762 ANNGYVSRKEHKSNTKQDDSEDIDEAKSEEMSDSEPESSKSEKKNKWKTEEVKKL 816
A NG V R+E+ +DD ED ++ K SDS + S+ E K +EE K+
Sbjct: 111 AKNGSVVRRENNGVGDEDDVEDEEQEKRALDSDSPKQESEIEDDCKEGSEEXXKI 275
>TC90087 similar to PIR|T05841|T05841 spliceosome-associated protein homolog
F17L22.120 - Arabidopsis thaliana, partial (12%)
Length = 733
Score = 33.5 bits (75), Expect = 0.36
Identities = 29/92 (31%), Positives = 42/92 (45%)
Frame = +2
Query: 725 VEIETITTAAEGDLSDSGESDEFWKPFIASSVEKSIKANNGYVSRKEHKSNTKQDDSEDI 784
VEIE + AE G +EF K F S + I + + N K+D+S +
Sbjct: 320 VEIEYVPEKAE---LYEGLDEEFKKIFEKFSFSEVI----------DSEDNDKKDESAEN 460
Query: 785 DEAKSEEMSDSEPESSKSEKKNKWKTEEVKKL 816
K + SDSE E +E+K K + + KKL
Sbjct: 461 AITKKKADSDSEEEEDDNEQKEKGVSNKKKKL 556
>TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity to
apoptosis antagonizing transcription
factor~gene_id:MFB13.10, partial (37%)
Length = 908
Score = 33.1 bits (74), Expect = 0.47
Identities = 23/97 (23%), Positives = 46/97 (46%)
Frame = +2
Query: 775 NTKQDDSEDIDEAKSEEMSDSEPESSKSEKKNKWKTEEVKKLIDLRSDLRDRFKVVKGRM 834
N DDSE +E + ++ E E + +++++WK +E+++L DL + + +
Sbjct: 101 NEVNDDSEVEEEEEEDDEGSDEEEEDERQEESRWKDDEMEQLEKEYMDLHHQEQELNTLK 280
Query: 835 ALWEEISQSLLADGISRSPGQCKSLWTSLALKYEVRF 871
L + LL ++ K+LW + E+RF
Sbjct: 281 NLKHHKDEDLLKGQAVKTQ---KALWYKI---LELRF 373
>TC81139 similar to PIR|D86427|D86427 hypothetical protein AAG30964.1
[imported] - Arabidopsis thaliana, partial (56%)
Length = 741
Score = 32.3 bits (72), Expect = 0.80
Identities = 27/93 (29%), Positives = 45/93 (48%), Gaps = 11/93 (11%)
Frame = +3
Query: 129 GNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSH----KIEALVITHGHEDHIGAL 184
G++ YILID G F + T +R + H KI+++++TH H D + L
Sbjct: 261 GSNHNYILIDVGKTFRE-------------TVLRWFVHHRIPKIDSIILTHEHADAVLGL 401
Query: 185 P---WVIPALDSN----TPIFASSFTMELIKKR 210
V P +N TPI+ S +M+ I+++
Sbjct: 402 DDVRAVQPFSPTNDIDPTPIYLSQHSMDSIEEK 500
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 32.0 bits (71), Expect = 1.0
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 7/93 (7%)
Frame = +1
Query: 726 EIETITTAAEGDLSDSGESDEFWKPFIASSVEKSIKANNGYVSRKEHKSNTKQDDS---- 781
E ET TT +E + E+D P +++EKS K + G + E ++ TK + +
Sbjct: 28 EKETTTTESETNTPTKSETDV---PETTTTIEKS-KNDLGTTTESETETPTKSETNVPET 195
Query: 782 ---EDIDEAKSEEMSDSEPESSKSEKKNKWKTE 811
E I + SEE +PE + K K + E
Sbjct: 196 TTTEPIVQTDSEETPTEKPEKETTTKTTKPEAE 294
>TC90136 weakly similar to PIR|T04494|T04494 hypothetical protein F8F16.90 -
Arabidopsis thaliana, partial (23%)
Length = 700
Score = 31.2 bits (69), Expect = 1.8
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +2
Query: 837 WEEISQSLLADGISRSPGQCKSLWTSLALKYE 868
W+ ISQ+ A + R+ QC+ W SL +YE
Sbjct: 170 WDIISQNCAALDVDRNLAQCRRKWRSLLAEYE 265
>TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown protein
{Arabidopsis thaliana}, partial (17%)
Length = 1237
Score = 30.8 bits (68), Expect = 2.3
Identities = 34/153 (22%), Positives = 65/153 (42%), Gaps = 1/153 (0%)
Frame = +2
Query: 666 AIENPGAVFADEINTKLSGKSQVGPGISTFRRSVDEHRKENQSTALQIRDDGIDIEGLLV 725
++ G + +TK S KS V + + + K+ +L D ++ G
Sbjct: 164 SVRRKGKKASSSKSTKPSKKSNVVSEKEAEKTADSKSSKKEVPISLN-EDSVVEATGTSE 340
Query: 726 EIETITTAAEGDLSDSGESDEFWKPFIASSVEKSIKANNGYVSRKEHKSNTKQDDSEDID 785
+ I + ESD P + S ++ V K++ S+ K+ +EDI
Sbjct: 341 NDKEIKAKISSPKAGGLESDAAGSPSPSESNHDENRSKKR-VRTKKNDSSAKEVAAEDIS 517
Query: 786 EAKSEEMSDSEPESSK-SEKKNKWKTEEVKKLI 817
+ SE SDS+ + ++ S KK ++ +VK ++
Sbjct: 518 KKVSEGTSDSKVKPARPSAKKGPIRSSDVKTVV 616
>BQ123319 similar to GP|10140692|gb| kinesin-like protein {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 788
Score = 30.4 bits (67), Expect = 3.0
Identities = 43/216 (19%), Positives = 79/216 (35%), Gaps = 20/216 (9%)
Frame = +3
Query: 620 LYKAAHAALSSCPVKSPLPHMERTVSEVLRKMVRKYSGKRPEVIAIAIENPG-------- 671
L K AA + +S + H TV+ V RK SG++ V + A EN G
Sbjct: 99 LEKELMAARNLANTRSAVTH---TVNGVHRKYNDPRSGRKARVSSRANENVGPGRDELES 269
Query: 672 -AVFADEINTKLSGKSQVGPGIST-----------FRRSVDEHRKENQSTALQIRDDGID 719
++ D++ +L + Q + R V+E +K S + + +
Sbjct: 270 WSLEVDDLKMELQARKQREAALEAALAEKEIMEEEHRNRVEEAKKRESSLENDLANMWVL 449
Query: 720 IEGLLVEIETITTAAEGDLSDSGESDEFWKPFIASSVEKSIKANNGYVSRKEHKSNTKQD 779
+ L E+ + + +S GE+ H ++ K +
Sbjct: 450 VAKLKKEVGVVAESNIDKISGDGEA---------------------------HTNDPKTN 548
Query: 780 DSEDIDEAKSEEMSDSEPESSKSEKKNKWKTEEVKK 815
DSE +K + + SEP + +++ W + KK
Sbjct: 549 DSESNIISKEQTLDVSEPNNETPKEEPFWLSSPGKK 656
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,227,274
Number of Sequences: 36976
Number of extensions: 295329
Number of successful extensions: 1640
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1622
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1635
length of query: 871
length of database: 9,014,727
effective HSP length: 105
effective length of query: 766
effective length of database: 5,132,247
effective search space: 3931301202
effective search space used: 3931301202
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC126782.9


