

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.17 - phase: 0 /pseudo
(346 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
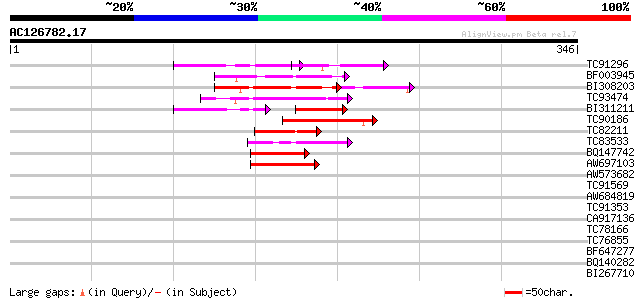
Score E
Sequences producing significant alignments: (bits) Value
TC91296 similar to PIR|T01223|T01223 hypothetical protein F6N23.... 43 2e-09
BF003945 52 3e-07
BI308203 GP|16508160|gb phosphoinositide-specific phospholipase ... 51 6e-07
TC93474 GP|23615322|emb|CAD52313. hypothetical protein {Plasmodi... 49 3e-06
BI311211 39 5e-06
TC90186 homologue to PIR|T03740|T03740 glucose-6-phosphate 1-deh... 43 2e-04
TC82211 homologue to GP|5566266|gb|AAD45353.1| glycinamide ribon... 42 3e-04
TC83533 42 5e-04
BQ147742 similar to PIR|C96525|C965 protein T1N15.17 [imported] ... 41 6e-04
AW697103 41 8e-04
AW573682 similar to GP|7940277|gb|A F2401.8 {Arabidopsis thalian... 39 0.002
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 39 0.003
AW684819 weakly similar to GP|21593795|gb| coclaurine N-methyltr... 33 0.003
TC91353 similar to GP|8777421|dbj|BAA97011.1 gene_id:K9P8.4~pir|... 39 0.004
CA917136 GP|13272475|gb unknown protein {Arabidopsis thaliana}, ... 38 0.005
TC78166 similar to GP|3075391|gb|AAC14523.1| unknown protein {Ar... 37 0.015
TC76855 similar to PIR|H96729|H96729 probable alanine aminotrans... 28 0.024
BF647277 similar to GP|15810008|gb| At1g63980/F22C12_9 {Arabidop... 35 0.024
BQ140282 similar to GP|15810549|gb| putative caffeoyl-CoA O-meth... 35 0.033
BI267710 similar to GP|12321970|gb unknown protein; 24137-33208 ... 34 0.073
>TC91296 similar to PIR|T01223|T01223 hypothetical protein F6N23.5 -
Arabidopsis thaliana, partial (7%)
Length = 629
Score = 42.7 bits (99), Expect(2) = 2e-09
Identities = 36/80 (45%), Positives = 43/80 (53%)
Frame = +2
Query: 101 SLMLCLIREIQTLENKWISLSKVVMIATWKLL**HKQL*EFMKTTYQYDMFISCFQLIFI 160
SL+ C R TL I + V +I +L FMKTTY +F S F IFI
Sbjct: 248 SLLRCCCRRHSTLGGISIHSNNVSLIVIRRLC-----SGRFMKTTYI--IFKSFF*RIFI 406
Query: 161 SSLR*LM*TTYSSYKKL*LY 180
SSL *L+ TTYS YKKL L+
Sbjct: 407 SSLL*LIKTTYSIYKKLTLF 466
Score = 36.6 bits (83), Expect(2) = 2e-09
Identities = 27/63 (42%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Frame = +3
Query: 173 SYKKL*LYFILCYRNSL----YKCLPYDSVYTMNCTLSCLSTCAKSSFFSKVHLHL*TLH 228
+Y K *LYF+LCYRNSL +KCL Y +S LS A F S + L L* +
Sbjct: 441 AYTKN*LYFVLCYRNSLCELIHKCLFY------KLQISSLSK*APKMFASCIFLLL*LIL 602
Query: 229 CLV 231
L+
Sbjct: 603 ILL 611
>BF003945
Length = 607
Score = 52.0 bits (123), Expect = 3e-07
Identities = 38/84 (45%), Positives = 50/84 (59%), Gaps = 2/84 (2%)
Frame = +2
Query: 126 IATWKLL**HKQ--L*EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYFIL 183
I ++L+**HK + +KT Y+YD F QLIFIS L *L+ TYS+ K +* YFI
Sbjct: 2 IYLFELI**HKLRLFGKLLKTAYEYDNF----QLIFISFLI*LIKKTYST-KTI*CYFIF 166
Query: 184 CYRNSLYKCLPYDSVYTMNCTLSC 207
CY+N L K + + C LSC
Sbjct: 167 CYKNDLSKLITIRA-----CALSC 223
>BI308203 GP|16508160|gb phosphoinositide-specific phospholipase C {Medicago
truncatula}, partial (5%)
Length = 811
Score = 51.2 bits (121), Expect = 6e-07
Identities = 48/136 (35%), Positives = 73/136 (53%), Gaps = 14/136 (10%)
Frame = +3
Query: 126 IATWKLL**HKQL*----EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYF 181
I+ ++L+ HK L* + +KTTY DM +SCFQL SSLR*+ T Y +++ L F
Sbjct: 399 ISLFELIYRHKNL*VCL*KLIKTTY--DMPMSCFQLNSKSSLR*VKKTAYK--QRILLCF 566
Query: 182 ILCYRNSLYKCLPYDSVYTMNCTLSCLSTCAKSSFFSKVHLHL-*TLHCLVSHLNILFIS 240
ILCY N+LY+ + Y ++ LS + T F H ++ L C + +NI+ +
Sbjct: 567 ILCY*NNLYRSTYMLNAYVIST*LSFIQTV----MFQL*HPYIFLMLPCSLIDMNIISDN 734
Query: 241 Y---------YLINTY 247
+ YLIN+Y
Sbjct: 735 FPSVLLSFLLYLINSY 782
Score = 47.0 bits (110), Expect = 1e-05
Identities = 36/81 (44%), Positives = 50/81 (61%), Gaps = 4/81 (4%)
Frame = +3
Query: 126 IATWKLL**HKQL*----EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYF 181
I+ ++L+ HK L* + +KTTY DM +SCFQL SSLR*+ T Y +++ L F
Sbjct: 144 ISLFELIYRHKNL*VCL*KLIKTTY--DMPMSCFQLNSKSSLR*VKKTAYK--QRILLCF 311
Query: 182 ILCYRNSLYKCLPYDSVYTMN 202
ILCY N+LY+ S Y +N
Sbjct: 312 ILCY*NNLYR-----STYMLN 359
>TC93474 GP|23615322|emb|CAD52313. hypothetical protein {Plasmodium
falciparum 3D7}, partial (2%)
Length = 811
Score = 48.9 bits (115), Expect = 3e-06
Identities = 40/97 (41%), Positives = 52/97 (53%), Gaps = 4/97 (4%)
Frame = +3
Query: 117 WISLSKVVMIATWKLL**HK----QL*EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYS 172
WI S+++ HK +L E +KT Y +F CFQ IF+SS R*LM
Sbjct: 66 WIDFSELIYY--------HKHLGDRLVELIKTVY--GLFRGCFQPIFVSSSR*LMKKQIV 215
Query: 173 SYKKL*LYFILCYRNSLYKCLPYDSVYTMNCTLSCLS 209
K +*LYFI CYRN+L L S Y+++ LS LS
Sbjct: 216 FMKIV*LYFISCYRNNL*VFL-MRSAYSISFKLSNLS 323
>BI311211
Length = 616
Score = 38.5 bits (88), Expect(2) = 5e-06
Identities = 18/32 (56%), Positives = 22/32 (68%)
Frame = -3
Query: 175 KKL*LYFILCYRNSLYKCLPYDSVYTMNCTLS 206
K +*LY I CYRNSLYK + S Y ++C LS
Sbjct: 416 KII*LYVIFCYRNSLYKFIIMISAYAISCKLS 321
Score = 28.9 bits (63), Expect(2) = 5e-06
Identities = 23/59 (38%), Positives = 30/59 (49%)
Frame = -1
Query: 101 SLMLCLIREIQTLENKWISLSKVVMIATWKLL**HKQL*EFMKTTYQYDMFISCFQLIF 159
S L L ++ T + W +L K M L * MKTTY ++FISCF+LIF
Sbjct: 613 STYLSLYTDMNTCDTVWTNL*KQFMYL*DCLD*-------LMKTTY--NIFISCFELIF 464
Score = 27.3 bits (59), Expect = 8.9
Identities = 16/29 (55%), Positives = 18/29 (61%)
Frame = -2
Query: 157 LIFISSLR*LM*TTYSSYKKL*LYFILCY 185
L FI+S +* M*TTYS YK LCY
Sbjct: 471 LFFINSSQ*FM*TTYSLYKN---NLTLCY 394
>TC90186 homologue to PIR|T03740|T03740 glucose-6-phosphate 1-dehydrogenase
(EC 1.1.1.49) TPG18 - common tobacco, partial (38%)
Length = 1168
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 4/62 (6%)
Frame = +3
Query: 167 M*TTYSSYKKL-*LYFILCYRNSLYKCLPYDSVYTMNCTLSCLSTCAKS---SFFSKVHL 222
+*+ + +Y ++ *LYFI CYRNSLY VY ++ LSCLST S SF S+V +
Sbjct: 27 L*SIFMAYMRIV*LYFIFCYRNSLYISTYMIDVYAIST*LSCLSTQRLSP*LSFASRVEV 206
Query: 223 HL 224
L
Sbjct: 207 GL 212
>TC82211 homologue to GP|5566266|gb|AAD45353.1| glycinamide ribonucleotide
transformylase {Vigna unguiculata}, partial (61%)
Length = 854
Score = 42.0 bits (97), Expect = 3e-04
Identities = 25/41 (60%), Positives = 29/41 (69%)
Frame = +1
Query: 150 MFISCFQLIFISSLR*LM*TTYSSYKKL*LYFILCYRNSLY 190
+FIS FQLI ISS LM*TTY Y +* YFILCY +L+
Sbjct: 733 LFISWFQLISISSSGQLM*TTYRLY-TV*FYFILCYTYNLF 852
>TC83533
Length = 692
Score = 41.6 bits (96), Expect = 5e-04
Identities = 28/64 (43%), Positives = 38/64 (58%)
Frame = +3
Query: 146 YQYDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYFILCYRNSLYKCLPYDSVYTMNCTL 205
Y + +F + F +F *LM T Y K +*LYF +CYRNSLY+ + S Y ++ L
Sbjct: 411 YVHKLFSAYFFKLFKI---*LMRTAYM--KTI*LYFTVCYRNSLYRSIYMISAYGISN*L 575
Query: 206 SCLS 209
SCLS
Sbjct: 576 SCLS 587
Score = 28.9 bits (63), Expect = 3.0
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = +1
Query: 148 YDMFISCFQLIFISSLR 164
YDM ISCFQLI +SS R
Sbjct: 406 YDMSISCFQLISLSSSR 456
>BQ147742 similar to PIR|C96525|C965 protein T1N15.17 [imported] -
Arabidopsis thaliana, partial (11%)
Length = 551
Score = 41.2 bits (95), Expect = 6e-04
Identities = 25/37 (67%), Positives = 28/37 (75%), Gaps = 1/37 (2%)
Frame = -1
Query: 148 YDMFISCFQLIFISSLR*LM*TTYSSYK-KL*LYFIL 183
YDMFI FQLI ISSLR*LM TTY+ Y+ L L+ IL
Sbjct: 437 YDMFICYFQLISISSLR*LMKTTYNFYENNLTLFHIL 327
Score = 29.3 bits (64), Expect = 2.3
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = -2
Query: 172 SSYKKL*LYFILCYRNSLYKCLPYD 196
+S K +*LYFI C+RNSL YD
Sbjct: 364 TSMKTI*LYFIFCFRNSLIHKH*YD 290
>AW697103
Length = 649
Score = 40.8 bits (94), Expect = 8e-04
Identities = 22/42 (52%), Positives = 26/42 (61%)
Frame = -2
Query: 148 YDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYFILCYRNSL 189
YD+ F FI S +*LM TTYS + +*LY I CY NSL
Sbjct: 519 YDIVHELFSDYFIKSQK*LMKTTYSLHDTI*LYIIFCYINSL 394
>AW573682 similar to GP|7940277|gb|A F2401.8 {Arabidopsis thaliana}, partial
(25%)
Length = 738
Score = 39.3 bits (90), Expect = 0.002
Identities = 25/74 (33%), Positives = 39/74 (51%)
Frame = -1
Query: 175 KKL*LYFILCYRNSLYKCLPYDSVYTMNCTLSCLSTCAKSSFFSKVHLHL*TLHCLVSHL 234
K +*LYFIL YRNSL+ S+Y ++C ++CLS + + TL L
Sbjct: 666 KSV*LYFILYYRNSLFITTYLISMYAISCKITCLSKQRPTVY---------TLFT*TG*L 514
Query: 235 NILFISYYLINTYI 248
++ I Y ++TY+
Sbjct: 513 SVYNICTYHLSTYL 472
Score = 32.0 bits (71), Expect = 0.36
Identities = 31/95 (32%), Positives = 47/95 (48%), Gaps = 5/95 (5%)
Frame = -3
Query: 153 SCFQLIFISSLR*LM*TTYSSYK-KL*LYFILCYRNSLYKCLPYDS--VYTM--NCTLSC 207
SCFQLIFISS * TT Y+ L L++++ + ++ L +D Y + N +
Sbjct: 733 SCFQLIFISSSG*YNETTNRLYEISLTLFYLILQK*LIHNHL-FDKHVCYKLQNNLFIKT 557
Query: 208 LSTCAKSSFFSKVHLHL*TLHCLVSHLNILFISYY 242
C + +++ L* +H H LFISYY
Sbjct: 556 TPNCLYPFYINRLIKCL*HMHLSFKH---LFISYY 461
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 38.9 bits (89), Expect = 0.003
Identities = 22/34 (64%), Positives = 26/34 (75%)
Frame = -1
Query: 142 MKTTYQYDMFISCFQLIFISSLR*LM*TTYSSYK 175
MKTTY +MFIS FQ IFI+SL+ L+* YS YK
Sbjct: 1290 MKTTY--NMFISYFQFIFINSLKQLL*IAYS*YK 1195
>AW684819 weakly similar to GP|21593795|gb| coclaurine N-methyltransferase
{Arabidopsis thaliana}, partial (24%)
Length = 666
Score = 33.5 bits (75), Expect(2) = 0.003
Identities = 21/35 (60%), Positives = 24/35 (68%)
Frame = -2
Query: 140 EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYSSY 174
E MKTTY DM I+ +QLI ISS *LM T Y+ Y
Sbjct: 473 ELMKTTY--DMSINRYQLISISST**LMKTVYNLY 375
Score = 24.3 bits (51), Expect(2) = 0.003
Identities = 10/28 (35%), Positives = 14/28 (49%)
Frame = -1
Query: 180 YFILCYRNSLYKCLPYDSVYTMNCTLSC 207
+ + RN+LYK Y+SV C C
Sbjct: 360 FILSLIRNNLYKSFLYESVLKNTCI*DC 277
>TC91353 similar to GP|8777421|dbj|BAA97011.1
gene_id:K9P8.4~pir||T04010~strong similarity to unknown
protein {Arabidopsis thaliana}, partial (11%)
Length = 855
Score = 38.5 bits (88), Expect = 0.004
Identities = 25/60 (41%), Positives = 33/60 (54%), Gaps = 1/60 (1%)
Frame = +1
Query: 151 FISCFQLIFISSLR*LM*TTYSSY-KKL*LYFILCYRNSLYKCLPYDSVYTMNCTLSCLS 209
F++C IFISS R* M T Y+ Y L L+ +L ++ + KC Y M LSCLS
Sbjct: 604 FMACS*FIFISSPR*FMKTAYNLYINNLTLFCLLLWK*LIRKCFIMI*AYAMRFKLSCLS 783
>CA917136 GP|13272475|gb unknown protein {Arabidopsis thaliana}, partial (7%)
Length = 779
Score = 38.1 bits (87), Expect = 0.005
Identities = 29/73 (39%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Frame = -1
Query: 135 HKQL*EFMKTTYQ--YDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYFIL------CYR 186
HK+ *+ +K + YDM ISCF I ISS L+ T YS Y F L ++
Sbjct: 620 HKRQ*DKLKKLMEIAYDMSISCFHHIPISSQGWLIKTAYSIYGNGSTLFYLLL*K*FIHK 441
Query: 187 NSLYKCLPYDSVY 199
+S YKC +DS Y
Sbjct: 440 HSYYKCQYHDSPY 402
Score = 28.5 bits (62), Expect = 4.0
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = -2
Query: 179 LYFILCYRNSLYKCLPYDSVYTM 201
LYFI C RNSLY P SV M
Sbjct: 481 LYFIFCCRNSLYISTPIISVNIM 413
>TC78166 similar to GP|3075391|gb|AAC14523.1| unknown protein {Arabidopsis
thaliana}, partial (13%)
Length = 1511
Score = 36.6 bits (83), Expect = 0.015
Identities = 36/88 (40%), Positives = 49/88 (54%), Gaps = 2/88 (2%)
Frame = -1
Query: 124 VMIATWKLL**HKQL*EFMKTTYQ--YDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYF 181
V I ++++**+ L*+ ++ + YDM IS FQLI I+ *L+*T S * F
Sbjct: 632 VWIDLFEVI**YNHL*DCLEELIKITYDMAISYFQLISINCS**LI*TINS----**TSF 465
Query: 182 ILCYRNSLYKCLPYDSVYTMNCTLSCLS 209
L RNSL Y +Y M+ LSCLS
Sbjct: 464 YLWLRNSL-----YIGMYAMSA*LSCLS 396
>TC76855 similar to PIR|H96729|H96729 probable alanine aminotransferase
F5A18.24 - Arabidopsis thaliana, partial (21%)
Length = 988
Score = 28.1 bits (61), Expect(2) = 0.024
Identities = 15/29 (51%), Positives = 17/29 (57%)
Frame = +3
Query: 161 SSLR*LM*TTYSSYKKL*LYFILCYRNSL 189
SS+ * M TY+ YK FI CYRN L
Sbjct: 555 SSIS*HMKITYNLYKNNLTLFIFCYRNRL 641
Score = 26.6 bits (57), Expect(2) = 0.024
Identities = 13/15 (86%), Positives = 13/15 (86%)
Frame = +2
Query: 148 YDMFISCFQLIFISS 162
YDM IS FQLIFISS
Sbjct: 503 YDMSIS*FQLIFISS 547
>BF647277 similar to GP|15810008|gb| At1g63980/F22C12_9 {Arabidopsis
thaliana}, partial (9%)
Length = 671
Score = 35.4 bits (80), Expect(2) = 0.024
Identities = 25/54 (46%), Positives = 31/54 (57%)
Frame = +2
Query: 140 EFMKTTYQYDMFISCFQLIFISSLR*LM*TTYSSYKKL*LYFILCYRNSLYKCL 193
E MKT Y DMFI CFQL +IS *LM T Y + L+ IL + ++K L
Sbjct: 431 ELMKTIY--DMFICCFQLTYISFPI*LMKTAYRT-----LFNILL*KKPIHKYL 571
Score = 29.6 bits (65), Expect = 1.8
Identities = 35/105 (33%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Frame = +3
Query: 100 FSLMLCLIREIQTLENKWISLSKVVMIATWKLL**HKQL*-EFMKTTYQ-YDMFISCFQL 157
+S+MLC + L WISL +++ ++ + +* + L* +FM +Y + + I FQ
Sbjct: 333 YSIMLCYVT--LCLGRVWISLVELMHVSICETV*--QNL*KQFMTCSYAVFSLPI*VFQ- 497
Query: 158 IFISSLR*LM*TTYSSYKKL*-LYFILCYRNSLY------KCLPY 195
YS K+L LY I CYR SLY KCL Y
Sbjct: 498 -------------YSL*KQLIELYLIFCYRKSLYISTYHDKCLCY 593
Score = 19.2 bits (38), Expect(2) = 0.024
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = +1
Query: 197 SVYTMNCTLSCLSTCAKSSF 216
SVY ++ LSCLS ++ F
Sbjct: 580 SVYAIS*KLSCLSKESEFGF 639
>BQ140282 similar to GP|15810549|gb| putative caffeoyl-CoA
O-methyltransferase {Arabidopsis thaliana}, partial (6%)
Length = 689
Score = 35.4 bits (80), Expect = 0.033
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 6/53 (11%)
Frame = +2
Query: 143 KTTYQYDMFISCFQ------LIFISSLR*LM*TTYSSYKKL*LYFILCYRNSL 189
KT + +FI C L+ + L LM TTY Y+K+*LY + C+RNSL
Sbjct: 74 KT*ACFFLFIICLD*LI*AYLLA*TLLXELMKTTYDMYEKI*LYALFCFRNSL 232
>BI267710 similar to GP|12321970|gb unknown protein; 24137-33208 {Arabidopsis
thaliana}, partial (2%)
Length = 591
Score = 34.3 bits (77), Expect = 0.073
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = -3
Query: 175 KKL*LYFILCYRNSLYKCLPYDSVYTMNCTLSCL 208
K + LYF+ CY+NSLY +D ++ LSCL
Sbjct: 577 KPIXLYFVFCYKNSLYISANHDKYLCVS*KLSCL 476
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.357 0.157 0.562
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,565,031
Number of Sequences: 36976
Number of extensions: 265817
Number of successful extensions: 3845
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 2858
Number of HSP's successfully gapped in prelim test: 135
Number of HSP's that attempted gapping in prelim test: 909
Number of HSP's gapped (non-prelim): 3090
length of query: 346
length of database: 9,014,727
effective HSP length: 97
effective length of query: 249
effective length of database: 5,428,055
effective search space: 1351585695
effective search space used: 1351585695
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC126782.17


