

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.15 + phase: 0
(1155 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
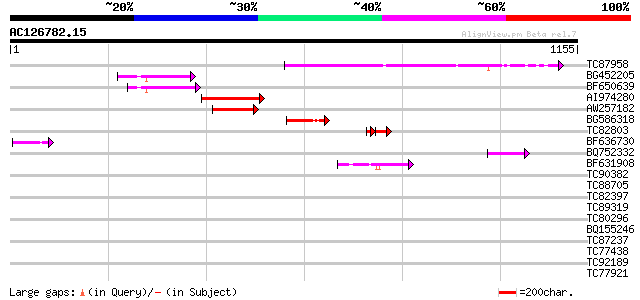
Sequences producing significant alignments: (bits) Value
TC87958 weakly similar to GP|20279447|gb|AAM18727.1 putative TNP... 249 3e-66
BG452205 weakly similar to GP|1345502|emb TNP2 {Antirrhinum maju... 133 5e-31
BF650639 weakly similar to GP|22748425|g putative transposon pro... 129 8e-30
AI974280 weakly similar to GP|20279447|g putative TNP2-like tran... 123 4e-28
AW257182 weakly similar to GP|20279447|g putative TNP2-like tran... 100 6e-21
BG586318 similar to GP|6069576|dbj| transposon-like ORF {Brassic... 83 5e-16
TC82803 similar to PIR|D85049|D85049 probable transposon protein... 57 5e-11
BF636730 62 2e-09
BQ752332 weakly similar to GP|8778366|gb| F15O4.30 {Arabidopsis ... 59 8e-09
BF631908 weakly similar to GP|21450441|gb| putative transposon p... 55 1e-07
TC90382 similar to GP|20161447|dbj|BAB90371. hypothetical protei... 35 0.17
TC88705 similar to GP|10177834|dbj|BAB11263. gene_id:MCD7.9~unkn... 33 0.83
TC82397 similar to GP|18377877|gb|AAL67124.1 AT3g53630/F4P12_330... 32 1.4
TC89319 similar to PIR|T02350|T02350 hypothetical protein T8F5.5... 32 1.8
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 32 1.8
BQ155246 32 1.8
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 32 1.8
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 31 2.4
TC92189 weakly similar to GP|21536818|gb|AAM61150.1 F-box protei... 31 2.4
TC77921 similar to GP|21553837|gb|AAM62930.1 phytochelatin synth... 31 3.1
>TC87958 weakly similar to GP|20279447|gb|AAM18727.1 putative TNP2-like
transposon protein {Oryza sativa (japonica
cultivar-group)}, partial (10%)
Length = 2433
Score = 249 bits (637), Expect = 3e-66
Identities = 186/593 (31%), Positives = 293/593 (49%), Gaps = 25/593 (4%)
Frame = +1
Query: 561 DLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELK 620
+LPYW N L HNLDVMHIEKN+FDNI T++D+ GKTKD+ + R DL+ RK+L +
Sbjct: 1 ELPYWASNTLHHNLDVMHIEKNIFDNIIGTLLDIPGKTKDHVRGRYDLEEMGIRKNLHPR 180
Query: 621 PQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDS 680
G++ + ++++++E + C LK ++PDG +SNI RC V+ ++ G KSHD+
Sbjct: 181 DVGGGRVAIANSCFSMTSEEKSMFCGVLKGAKLPDGSASNISRCVKVSEKKICGYKSHDA 360
Query: 681 HIFMENFIPIAF-SSLPKHVLDPLIEISQFFKNLCASTLRVDELEKME-KNMPIIICQLE 738
H + + I S++PK V PLI +S+FF++LC ++ +N +
Sbjct: 361 HFMLHYLLQIPIRSTMPKAVAQPLIRLSRFFRSLCQKGYPNP*SGQLTGRNC*STLFN*R 540
Query: 739 QIFPPGF----FDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGS 794
F F F + + L + A +RWMYP ER++ K V+N ++ EGS
Sbjct: 541 WFFLLVFLMLWFT*LFIFRMRLDWVA----QFNFRWMYPVERYLCKLKGYVRNRSRPEGS 708
Query: 795 IVAIYTAKETTYFIGHYFVDRLLTPSNTRNE-VQVNDLRDPSTLSVFNLDGRHAG--KVL 851
I Y A+E Y + T SN +N+ +DL + + F+ GR G K
Sbjct: 709 IAEGYVAEEGITICSRYMHKGVDTRSNRKNQNYDDSDLLEVDEIDYFSSIGRPLGGKKNG 888
Query: 852 QYWMVDQKEMRSAHVHVLINCAEVKPYLEE---FIAFYAEFGEGSTVGSIHEYFPAWFKQ 908
+ + +D + AH ++L NC E++ Y+ E + + + + + F WFK
Sbjct: 889 KPFYLDFDSLAQAHRYLLFNCGEIEVYIREHEGIVNSHTKKRKWDKAKAQCLNFSDWFKT 1068
Query: 909 RVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT 968
R A+ D +LR+ S+GP A F Y VNGY+FHT +KT NSGV + +T
Sbjct: 1069 R---AKRDDVPPQLREFSRGPNNVAKRFSGYLVNGYRFHTMQ*DARRKTQNSGVTLVSLT 1239
Query: 969 ---DGGEDD--------FYGVIKHIYELSYNDN-KVVLFYCDWFDPSPRWTKINKLCNTV 1016
+D+ FYG I I EL+Y + K VLF CDW++ + C
Sbjct: 1240 ASFASSKDENPRTEPVNFYGAINDIIELNYYGHCKFVLFRCDWYEVEE--DEYGLTC--- 1404
Query: 1017 DIRVDKKYKEYDPFIMAHNVRQVYYVPYPPTLPRKRGWSVAIKTKPRGRIETEERNEEVA 1076
+ +KK + D F++A V+Q +YV P + R A+KT PR + ++E
Sbjct: 1405 -VYFNKKCYQDDAFVIASQVQQCFYVQDPLNVNR----HYALKTDPRDLFKMGNQSE--- 1560
Query: 1077 YQADDMTHVNEIIEVDQVTSWQDSQVEGDQVD-PSILEMRNEVEDENEDVVDD 1128
++A + +EI +W + ++ PS + E EDE+ DD
Sbjct: 1561 FEATSVNAADEI-------NWGREDIPRTIIEKPSNILQ*IEEEDEDYSDFDD 1698
>BG452205 weakly similar to GP|1345502|emb TNP2 {Antirrhinum majus}, partial
(16%)
Length = 619
Score = 133 bits (334), Expect = 5e-31
Identities = 69/172 (40%), Positives = 102/172 (59%), Gaps = 13/172 (7%)
Frame = +1
Query: 220 LPMNYHAAKRLVMKLGLSVKKIDCCRNGCMLFY-DNEFGINDGPLEECKFCQSPRYGIS- 277
+P +Y+ + ++ LGL KID C N CML++ D++ I+ C C + R+ S
Sbjct: 121 VPDSYNKTR*MIRDLGLDYSKIDACPNDCMLYWKDHKNDIS------CHVCGASRWIEST 282
Query: 278 ----------KQKRVAVKSMFYLPIIPRLQRLFASMKTASQMTWHHSN-GISGVMRHPSD 326
K+ +V+ K + + P+IPRLQRLF +TA+ + WHH + G +RHP+D
Sbjct: 283 TESGQIEENKKEHKVSTKILRHFPLIPRLQRLFMCSETATSLRWHHDDCSKDGKLRHPAD 462
Query: 327 GEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQSSAKPYSCWPIIVTPYN 378
G+AWK F+ HP+FA D RN+RLGL SDGF P+ + K Y WP+ + PYN
Sbjct: 463 GQAWKDFNGHHPEFARDSRNIRLGLASDGFNPFRAMNLK-YXAWPVXLIPYN 615
>BF650639 weakly similar to GP|22748425|g putative transposon protein {Oryza
sativa (japonica cultivar-group)}, partial (6%)
Length = 568
Score = 129 bits (323), Expect = 8e-30
Identities = 68/163 (41%), Positives = 94/163 (56%), Gaps = 13/163 (7%)
Frame = +2
Query: 240 KIDCCRNGCMLFY-DNEFGINDGPLEECKFCQSPRYGIS-----------KQKRVAVKSM 287
KID C N CML++ D++ I+ C C + R S K+ +V+ K +
Sbjct: 71 KIDACXNDCMLYWKDHKNDIS------CHVCGASRXIESTTEXGQIEENKKEHKVSTKIL 232
Query: 288 FYLPIIPRLQRLFASMKTASQMTWHHSNGIS-GVMRHPSDGEAWKHFDRVHPDFAADPRN 346
+ P+IP LQR F +TA+ + WHH + G +RHP+DG+AWK F+ HP+FA D RN
Sbjct: 233 RHFPLIPXLQRXFMCSETATSLRWHHDDXSKDGKLRHPADGQAWKDFNGHHPEFARDSRN 412
Query: 347 VRLGLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPY 389
+ LGL SDGF P+ + K YS WP I+ PYN PP +CM Y
Sbjct: 413 IXLGLASDGFNPFRAMNLK-YSAWPXILIPYNFPPWLCMKAXY 538
>AI974280 weakly similar to GP|20279447|g putative TNP2-like transposon
protein {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 457
Score = 123 bits (309), Expect = 4e-28
Identities = 63/128 (49%), Positives = 82/128 (63%), Gaps = 1/128 (0%)
Frame = +1
Query: 392 LSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAALMWTINDFPA 450
LS ++PGP +PK IDV+L+PLID+LK LW G YD S KQ F+ AAL+WTI+D PA
Sbjct: 16 LSLLIPGPSSPKRNIDVYLEPLIDELKELWERGFEVYDASSKQLFEAHAALLWTISDLPA 195
Query: 451 YGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDE 510
Y MLSGWST G+ ACP C +T + L+ K+ + HR FL H +R+ K GF +
Sbjct: 196 YTMLSGWSTSGKLACPVCSYDTCSMYLKHSRKTCYMS-HRRFLDPKHRWRKDKKGFDGKK 372
Query: 511 RVFVGPPP 518
+ V P P
Sbjct: 373 EIRVAPFP 396
>AW257182 weakly similar to GP|20279447|g putative TNP2-like transposon
protein {Oryza sativa (japonica cultivar-group)},
partial (6%)
Length = 387
Score = 99.8 bits (247), Expect = 6e-21
Identities = 49/94 (52%), Positives = 63/94 (66%), Gaps = 1/94 (1%)
Frame = +1
Query: 414 IDDLKRLWV-GELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENT 472
I++LK LW G TYD S+ Q F++RAAL+WTI+DFPAY MLSGWST G+ ACP C NT
Sbjct: 7 IEELKVLWESGVETYDASKNQMFRMRAALLWTISDFPAYAMLSGWSTKGKLACPCCNNNT 186
Query: 473 KAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGF 506
+ L+ K + D HR FLP +H +R + F
Sbjct: 187 SSTYLKHSRKMCYMD-HRTFLPMDHAWRENRKSF 285
>BG586318 similar to GP|6069576|dbj| transposon-like ORF {Brassica rapa},
partial (11%)
Length = 267
Score = 83.2 bits (204), Expect = 5e-16
Identities = 42/86 (48%), Positives = 56/86 (64%)
Frame = +3
Query: 565 WKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPN 624
W+D+LLRHNLDVMHIEKN FDN+ NT+++V+GKTKDN K+R DL C R +L + N
Sbjct: 3 WEDHLLRHNLDVMHIEKNFFDNLMNTILNVQGKTKDNLKSRLDLVDICARSELHV--DEN 176
Query: 625 GKLLKPKANYNLSAQEAKLVCQWLKD 650
G+ P Y L A + W+ +
Sbjct: 177 GR--APFPIYRLDAGGKDALFDWISN 248
>TC82803 similar to PIR|D85049|D85049 probable transposon protein [imported]
- Arabidopsis thaliana, partial (5%)
Length = 755
Score = 57.0 bits (136), Expect(2) = 5e-11
Identities = 25/33 (75%), Positives = 28/33 (84%)
Frame = +3
Query: 746 FDSMEHLPVHLAYEAILGGPVQYRWMYPFERFM 778
FDSMEHL +HLAYEA +G PVQYR MYPFER +
Sbjct: 288 FDSMEHLSIHLAYEARVGNPVQYR*MYPFERLI 386
Score = 29.6 bits (65), Expect(2) = 5e-11
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +1
Query: 728 KNMPIIICQLEQIFPPGF 745
+N+ II+C LEQIFPP F
Sbjct: 235 ENIAIIMC*LEQIFPPSF 288
>BF636730
Length = 637
Score = 61.6 bits (148), Expect = 2e-09
Identities = 30/83 (36%), Positives = 48/83 (57%)
Frame = +1
Query: 7 YRRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPG 66
+R+WM +R P + + +F+ GV F+ +A Q+ + G +RCPC C+CR +
Sbjct: 298 HRKWMDNRMRPDNSRVTDEFLAGVCEFVEFACAQDDFKKVGKLRCPCKICKCRKP*S-VD 474
Query: 67 DVKKHLKKVGFMPNYWVWTYNGE 89
DV KHL GF +Y+ W+ +GE
Sbjct: 475 DVMKHLCMKGFKEDYYYWSSHGE 543
>BQ752332 weakly similar to GP|8778366|gb| F15O4.30 {Arabidopsis thaliana},
partial (6%)
Length = 268
Score = 59.3 bits (142), Expect = 8e-09
Identities = 30/89 (33%), Positives = 45/89 (49%), Gaps = 3/89 (3%)
Frame = -2
Query: 974 DFYGVIKHIYELSYNDN---KVVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPF 1030
DFYG++ I E+ + K VLF C+WFDP + VD+ ++Y +++PF
Sbjct: 267 DFYGILTEIIEVEFPGILKLKCVLFKCEWFDPVVNRGVRSIKFGVVDVNGGRRYNKFEPF 88
Query: 1031 IMAHNVRQVYYVPYPPTLPRKRGWSVAIK 1059
I+A QV ++PYP W IK
Sbjct: 87 ILASQADQVSFLPYPRMRDSGINWLAVIK 1
>BF631908 weakly similar to GP|21450441|gb| putative transposon protein
5'-partial {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 681
Score = 55.5 bits (132), Expect = 1e-07
Identities = 51/169 (30%), Positives = 72/169 (42%), Gaps = 16/169 (9%)
Frame = +3
Query: 669 TGRLHGMKSHDSHIFMENFIPIAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKMEK 728
T +LH SH F + + + H ++ L E++ + L S + + +E+
Sbjct: 144 TVKLHRRISHRHSSFSPSTVTHS------HAVELLTELNLTVE-LHTSPIELHSRGSIER 302
Query: 729 NMPIIICQLEQIFPPGFF-----DSMEHL-----------PVHLAYEAILGGPVQYRWMY 772
+ PI QL+ F FF EHL + A+EA + GPV YRWMY
Sbjct: 303 HHPI--SQLQI*FHKSFFLFRSRRGREHL*GWIRAGYLRRDLQ*AHEARIAGPVHYRWMY 476
Query: 773 PFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSN 821
P ERF+ K V+N EGSI Y A E Y D + T N
Sbjct: 477 PIERFILTLKNYVRNRNYPEGSIAEGYVANECLTHCSRYMSDGVQTKFN 623
>TC90382 similar to GP|20161447|dbj|BAB90371. hypothetical protein~similar to
Arabidopsis thaliana chromosome 1 F8K7.2, partial (13%)
Length = 1047
Score = 35.0 bits (79), Expect = 0.17
Identities = 34/147 (23%), Positives = 64/147 (43%), Gaps = 3/147 (2%)
Frame = +3
Query: 1004 PRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYPPTLPRKRGWSVAIK--TK 1061
P +++ N+V ++ +KK + F+ RQ++ V P W +K TK
Sbjct: 156 PNSLELSVSINSVPMKEEKK--AFSSFVKKGG-RQMFTVELRPGETTIVSWKKLLKDATK 326
Query: 1062 PRGRIETEERNEEVAYQADDMTHVNEIIEVDQVTSWQDSQVEGDQVDPSILEMRN-EVED 1120
P G T +++ + +EV++ Q + L M N +D
Sbjct: 327 PNGSASTSQQSRPEPFPGQP-------VEVEENDPAQPHRFSAVIEKIERLYMGNGSSDD 485
Query: 1121 ENEDVVDDNNEGDHQDNQVDDGDVDEY 1147
E+ V D+++ D +D+ +DD ++DEY
Sbjct: 486 EDLPDVPDDDQYDTEDSFIDDAELDEY 566
>TC88705 similar to GP|10177834|dbj|BAB11263. gene_id:MCD7.9~unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1685
Score = 32.7 bits (73), Expect = 0.83
Identities = 28/107 (26%), Positives = 41/107 (38%), Gaps = 3/107 (2%)
Frame = +1
Query: 108 GTNVETNVEYNEDVNTDQFNMMDDM--VADALGVELSYGEDVEDDEEEQPPNEKAQRFYR 165
GTN E N Y + + D DD D G Y ED+ D+E ++ +E Y+
Sbjct: 601 GTNNEENEGYASETDDDTSKYDDDTGKYDDDTG---KYDEDINDEEFQEDEHEDLSSSYK 771
Query: 166 LLSESNTPLYEGSSH-SKLSMCVWLLSHKSNYLNPDDGMDDITKMLK 211
ES L + S K+ VW + N D ++ K
Sbjct: 772 SDVESEPDLSDDPSWLEKIQKSVWNIIQVVNIFQTPVNQSDAARIRK 912
>TC82397 similar to GP|18377877|gb|AAL67124.1 AT3g53630/F4P12_330 {Arabidopsis
thaliana}, partial (9%)
Length = 716
Score = 32.0 bits (71), Expect = 1.4
Identities = 32/132 (24%), Positives = 62/132 (46%), Gaps = 6/132 (4%)
Frame = +3
Query: 1021 DKKYKEYDPFIMAHNVRQVYYVPYPPTLPRKRGW-SVAIKTKPRGRIETEERNEEVAYQA 1079
+KK+K +A +V V+++ P T P K W V+IK + R++ + +
Sbjct: 132 EKKFKP-----IADHVNAVHHLHDPTTFPFKWSWRDVSIKVQ-------NMRHQYIGVKH 275
Query: 1080 DDMTHVNEIIEVDQVTSWQD----SQVEGD-QVDPSILEMRNEVEDENEDVVDDNNEGDH 1134
++ + D V W++ +V GD ++ P + ++ D E +DD+++ D
Sbjct: 276 KIQSNDHGFNWNDGVIHWENFLNYKRVFGDVKIHP-----KPKLNDGFETEIDDSDDSDG 440
Query: 1135 QDNQVDDGDVDE 1146
++ DGDV E
Sbjct: 441 DEDHDCDGDVSE 476
>TC89319 similar to PIR|T02350|T02350 hypothetical protein T8F5.5 -
Arabidopsis thaliana, partial (53%)
Length = 1436
Score = 31.6 bits (70), Expect = 1.8
Identities = 31/120 (25%), Positives = 53/120 (43%), Gaps = 6/120 (5%)
Frame = +1
Query: 509 DERVFVGPPPKITSEEVWT-----RVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLP 563
D +FVGPPP E + R + ++ + P GV+H+ + +I
Sbjct: 142 DTELFVGPPPPAMVSEAESANEAERFEEVTRIMEVEINSPYDVLGVNHNMSDGNI--KKK 315
Query: 564 YWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARK-DLQIYCKRKDLELKPQ 622
YWK +LL H H + N F + + +D EK ++ D +I K++ +LK +
Sbjct: 316 YWKISLLVHPDKCSHPQAN---QAFIKLNKAFKELQDPEKRKEMDEKIKLKQEQEDLKAE 486
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 31.6 bits (70), Expect = 1.8
Identities = 18/88 (20%), Positives = 38/88 (42%)
Frame = +3
Query: 1067 ETEERNEEVAYQADDMTHVNEIIEVDQVTSWQDSQVEGDQVDPSILEMRNEVEDENEDVV 1126
+ + +++V+ +DD + D S D + D V + ++ E +D
Sbjct: 804 DNAQEDDQVSLNSDDDNQLGS----DNTGSDDDEAEDHDGVSDDENDRSSDYETSGDDAD 971
Query: 1127 DDNNEGDHQDNQVDDGDVDEYNYASDNH 1154
+ +EGD ++ +DG + E+ D H
Sbjct: 972 NVEDEGDDLEDSEEDGGISEHEGDGDLH 1055
>BQ155246
Length = 387
Score = 31.6 bits (70), Expect = 1.8
Identities = 18/46 (39%), Positives = 25/46 (54%)
Frame = +3
Query: 972 EDDFYGVIKHIYELSYNDNKVVLFYCDWFDPSPRWTKINKLCNTVD 1017
ED FY I IY + Y +V + C++FD P+WT C +VD
Sbjct: 153 EDAFYFFIIIIYHVFY----LVNYKCNFFD*KPKWTS----CQSVD 266
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 31.6 bits (70), Expect = 1.8
Identities = 29/107 (27%), Positives = 43/107 (40%), Gaps = 11/107 (10%)
Frame = +1
Query: 1058 IKTKPRGRIETEERNEEVAYQADDMTHVNEIIEVDQVTSWQDSQVEGDQVDPSI------ 1111
+K G ET E EE + Q ++ T + + S Q+ +QV +
Sbjct: 292 LKKNEGGEKETGEITEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQ 471
Query: 1112 -LEMRNEVEDENEDVVDDNNEGDHQDNQVDDGDVD----EYNYASDN 1153
E N E ++ D E DNQV DG+ + E NY+ DN
Sbjct: 472 GTEETNGTEGGEKEEHDKIKEDTSSDNQVQDGEKNNEAREENYSGDN 612
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 - garden
pea, partial (93%)
Length = 1607
Score = 31.2 bits (69), Expect = 2.4
Identities = 19/82 (23%), Positives = 40/82 (48%), Gaps = 4/82 (4%)
Frame = +2
Query: 1069 EERNEEVAYQADDMTHVNEIIE---VDQVTSWQDSQV-EGDQVDPSILEMRNEVEDENED 1124
E+ EE+ Q + + I + SW + +G++ ++ +E +DE++D
Sbjct: 899 EDMAEELQNQMEQDYDIGSTIRDKIIPHAVSWFTGEAAQGEEFG----DLDDEDDDEDDD 1066
Query: 1125 VVDDNNEGDHQDNQVDDGDVDE 1146
DD+ + D +++ DD D +E
Sbjct: 1067 AEDDDEDEDEDEDEDDDEDEEE 1132
>TC92189 weakly similar to GP|21536818|gb|AAM61150.1 F-box protein family
AtFBW2 {Arabidopsis thaliana}, partial (32%)
Length = 846
Score = 31.2 bits (69), Expect = 2.4
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = +1
Query: 1102 VEGDQVDPSILEMRNEVEDENEDVVDDNNEGDHQDNQVDDGDVDEYNYASDN 1153
V Q P + + +V E + D ++ D D++ DD D+DEY+Y D+
Sbjct: 370 VSVQQKFPKLAVLGPQVIGYYEMLEDCSDISDSSDSEYDDSDMDEYDYDDDS 525
>TC77921 similar to GP|21553837|gb|AAM62930.1 phytochelatin synthetase-like
protein {Arabidopsis thaliana}, partial (82%)
Length = 1727
Score = 30.8 bits (68), Expect = 3.1
Identities = 14/45 (31%), Positives = 26/45 (57%)
Frame = +3
Query: 137 LGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSHS 181
+ + L GE + E + PN + +R +++ S ++T L EGS+ S
Sbjct: 360 MDIRLDMGEKGSNLEHDGKPNNRTRRLFKIQSRNSTLL*EGSNSS 494
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,366,075
Number of Sequences: 36976
Number of extensions: 704713
Number of successful extensions: 3472
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 3376
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3446
length of query: 1155
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1048
effective length of database: 5,058,295
effective search space: 5301093160
effective search space used: 5301093160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC126782.15


