

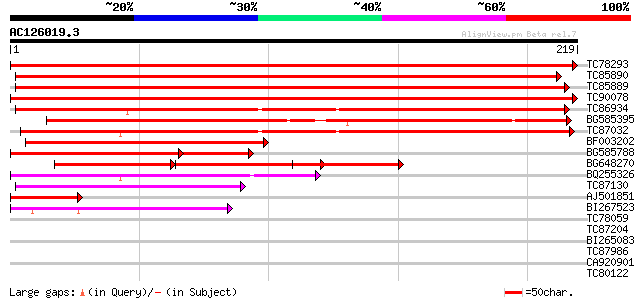
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.3 + phase: 0
(219 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78293 similar to SP|P47192|SYBR_ARATH Synaptobrevin-related pr... 436 e-123
TC85890 similar to SP|P47192|SYBR_ARATH Synaptobrevin-related pr... 352 6e-98
TC85889 similar to GP|22137292|gb|AAM91491.1 At1g04740/T1G11_1 {... 340 2e-94
TC90078 similar to GP|23617131|dbj|BAC20811. putative synaptobre... 329 4e-91
TC86934 similar to PIR|T04630|T04630 synaptobrevin homolog F10N7... 164 2e-41
BG585395 weakly similar to GP|19703107|gb synaptobrevin-like pro... 162 7e-41
TC87032 similar to GP|9757817|dbj|BAB08335.1 synaptobrevin-like ... 155 1e-38
BF003202 similar to PIR|F86180|F861 hypothetical protein [import... 137 3e-33
BG585788 homologue to GP|7268326|emb SYBL1 like protein {Arabido... 104 2e-23
BG648270 similar to GP|22137292|gb At1g04740/T1G11_1 {Arabidopsi... 76 9e-15
BQ255326 similar to GP|9757817|dbj synaptobrevin-like protein {A... 74 6e-14
TC87130 weakly similar to PIR|T04067|T04067 hypothetical protein... 54 4e-08
AJ501851 similar to PIR|F86180|F861 hypothetical protein [import... 48 3e-06
BI267523 42 2e-04
TC78059 similar to GP|4206787|gb|AAD11808.1| syntaxin-related pr... 32 0.25
TC87204 small G-protein ROP9 [Medicago truncatula] 31 0.42
BI265083 similar to GP|15450711|gb| At1g06210/F9P14_4 {Arabidops... 31 0.42
TC87986 similar to GP|5669636|gb|AAD46403.1| ethylene-responsive... 30 0.72
CA920901 homologue to GP|22597174|gb syntaxin {Glycine max}, par... 29 1.2
TC80122 similar to GP|19401700|gb|AAL87667.1 transcription facto... 28 2.7
>TC78293 similar to SP|P47192|SYBR_ARATH Synaptobrevin-related protein.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 1066
Score = 436 bits (1121), Expect = e-123
Identities = 219/219 (100%), Positives = 219/219 (100%)
Frame = +2
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD
Sbjct: 134 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 313
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY
Sbjct: 314 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 493
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG
Sbjct: 494 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 673
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC
Sbjct: 674 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 790
>TC85890 similar to SP|P47192|SYBR_ARATH Synaptobrevin-related protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (95%)
Length = 1087
Score = 352 bits (903), Expect = 6e-98
Identities = 168/211 (79%), Positives = 196/211 (92%)
Frame = +3
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNG 62
Q+SLIY+FV+RG+VIL+EY++F+GNF +IA QCLQKLPASNN+FTYNCDGHTF++LVDNG
Sbjct: 114 QKSLIYAFVSRGSVILSEYTEFSGNFNSIAFQCLQKLPASNNKFTYNCDGHTFNYLVDNG 293
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
+TYCVVA E+VGRQ+P+AFLER+KDDF +YGGG+A+TA SLNKEFGPKLKE MQYCV
Sbjct: 294 YTYCVVADETVGRQVPVAFLERVKDDFVAKYGGGKASTAAPNSLNKEFGPKLKEHMQYCV 473
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
+HPEEVSKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENL QAQDFR GT+
Sbjct: 474 DHPEEVSKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLHHQAQDFRNSGTK 653
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSV 213
+RRKMW QNMKIKLIVLAI+IALILIIVL +
Sbjct: 654 IRRKMWLQNMKIKLIVLAILIALILIIVLPI 746
>TC85889 similar to GP|22137292|gb|AAM91491.1 At1g04740/T1G11_1 {Arabidopsis
thaliana}, partial (94%)
Length = 992
Score = 340 bits (873), Expect = 2e-94
Identities = 160/214 (74%), Positives = 192/214 (88%)
Frame = +1
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNG 62
Q+ LIY+FV+RGTVILAE+++F+GNF +IA QCLQKLP++NN+FTYNCD HTF++L+DNG
Sbjct: 94 QKQLIYAFVSRGTVILAEFTEFSGNFNSIAFQCLQKLPSTNNKFTYNCDNHTFNYLIDNG 273
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
+TYCVVA E+ GRQ+P+AFLER+KDDF +YGG +A+TA SLNKEFGPKLKE MQYCV
Sbjct: 274 YTYCVVADETTGRQVPMAFLERVKDDFVSKYGGEKASTAPPNSLNKEFGPKLKEHMQYCV 453
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
+HP+E+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKT+NL QAQDFR GT
Sbjct: 454 DHPDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTDNLHHQAQDFRSSGTS 633
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
+RRKMW QNMK+KLIVL I+IALILIIVLSV G
Sbjct: 634 IRRKMWLQNMKVKLIVLGILIALILIIVLSVTRG 735
>TC90078 similar to GP|23617131|dbj|BAC20811. putative synaptobrevin {Oryza
sativa (japonica cultivar-group)}, partial (84%)
Length = 975
Score = 329 bits (844), Expect = 4e-91
Identities = 155/219 (70%), Positives = 188/219 (85%)
Frame = +2
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M Q+S IYSFVARGT++LAEY++FTGNF IA QCLQKLP+SNN+FTY+CD HTF+FLV+
Sbjct: 98 MSQESFIYSFVARGTMVLAEYTEFTGNFPAIAAQCLQKLPSSNNKFTYSCDHHTFNFLVE 277
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
+G+ YCVVA ESV +QI IAFLER+K DF KRYG G+A TA AKSLNKEFGP +KE M+Y
Sbjct: 278 DGYAYCVVAKESVSKQISIAFLERVKADFKKRYGVGKADTAIAKSLNKEFGPVMKEHMKY 457
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
++H EE+ KL KVKAQVSEVK +M+ENIDK IDRGE + VL DKTE LR+QAQDFR+QG
Sbjct: 458 IIDHAEEIEKLLKVKAQVSEVKSIMLENIDKAIDRGENLSVLSDKTETLRAQAQDFRKQG 637
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ+RRKMWYQNMKIKL+VL I++ L+L+I LS+C GF+C
Sbjct: 638 TQVRRKMWYQNMKIKLVVLGILLFLVLVIWLSICGGFNC 754
>TC86934 similar to PIR|T04630|T04630 synaptobrevin homolog F10N7.40 -
Arabidopsis thaliana, complete
Length = 1208
Score = 164 bits (416), Expect = 2e-41
Identities = 83/215 (38%), Positives = 137/215 (63%), Gaps = 1/215 (0%)
Frame = +2
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLVDN 61
+ ++Y+ VARGTV+LAE++ T N IA Q L+K+P +N+ +Y+ D + F +
Sbjct: 110 RMGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTD 289
Query: 62 GFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYC 121
G T +A +SVGR+IP AFLE I F + YG TA A ++N EF L +QM+Y
Sbjct: 290 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYF 466
Query: 122 VEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGT 181
P ++ ++K ++S+V+ VM+ENIDKV+DRG+++E+LVDK N++ FR+Q
Sbjct: 467 SSDPN-ADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQAR 643
Query: 182 QLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
+ R +W++N+K+ + ++ I+I + +++ VCHG
Sbjct: 644 RFRSTVWWRNVKLTIALIVILIVIAYVVLAFVCHG 748
>BG585395 weakly similar to GP|19703107|gb synaptobrevin-like protein
{Dictyostelium discoideum}, partial (44%)
Length = 668
Score = 162 bits (411), Expect = 7e-41
Identities = 88/206 (42%), Positives = 135/206 (64%), Gaps = 3/206 (1%)
Frame = +3
Query: 15 TVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFTYCVVAVESVG 74
+VILAEY++ +GNFT + L+K+P +N++ TY D + F ++ ++G TY +A +S G
Sbjct: 3 SVILAEYTNSSGNFTQVTQAILEKIPPNNSKLTYVYDRYLFHYICEDGLTYMCMADDSFG 182
Query: 75 RQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVEHPEEVS---KL 131
R+IP FL+ IK+ F Y RATTA +N EF +++QM EH +VS ++
Sbjct: 183 RRIPFLFLQDIKERFLVTYSKDRATTAVPYGMN-EFSKVIQKQM----EHFSDVSHADRI 347
Query: 132 AKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQN 191
KV ++ +VK VM NI++V++RGE+IE+LVDKT+NL QA F+++ T L+R MW++N
Sbjct: 348 KKVHGEIEQVKDVMTRNIEQVLERGERIEILVDKTDNLNQQAFAFKKRSTALKRSMWWKN 527
Query: 192 MKIKLIVLAIIIALILIIVLSVCHGF 217
K+ LI+L +I LI V+S GF
Sbjct: 528 TKL-LILLVFVIILITYFVISSACGF 602
>TC87032 similar to GP|9757817|dbj|BAB08335.1 synaptobrevin-like protein
{Arabidopsis thaliana}, partial (98%)
Length = 1153
Score = 155 bits (391), Expect = 1e-38
Identities = 78/215 (36%), Positives = 139/215 (64%), Gaps = 1/215 (0%)
Frame = +1
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA-SNNRFTYNCDGHTFSFLVDNGF 63
+++Y+ VARGTV+LAE+S TGN +A + L+KLP S++R ++ D + F L +G
Sbjct: 193 AILYALVARGTVVLAEFSAVTGNTGAVARRLLEKLPTESDSRLCFSQDRYIFHILRSDGL 372
Query: 64 TYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVE 123
T+ +A ++ GR+IP ++LE I+ F K Y A A A ++N EF L +QM++
Sbjct: 373 TFLCMANDTFGRRIPFSYLEDIQMRFMKNYSRV-ANYAPAYAMNDEFSRVLHQQMEFFSS 549
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
+P V L +V+ +V E++ +M++NI+K+++RG++IE+LVDKT ++ + FR+Q +L
Sbjct: 550 NPS-VDALNRVRGEVGEIRTIMVDNIEKILERGDRIELLVDKTATMQDSSFHFRKQSKRL 726
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
RR +W +N K+ ++ +I+ L+ ++ + C G +
Sbjct: 727 RRALWMKNFKLLALLTCLIVLLLYFLIAACCGGIT 831
>BF003202 similar to PIR|F86180|F861 hypothetical protein [imported] -
Arabidopsis thaliana, partial (41%)
Length = 325
Score = 137 bits (345), Expect = 3e-33
Identities = 61/94 (64%), Positives = 81/94 (85%)
Frame = +3
Query: 7 IYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFTYC 66
IY+FV+RGTVILAE+++ + +IALQ LQKLP++NN+FTYNCD HTF++L+DNG+TYC
Sbjct: 3 IYAFVSRGTVILAEFTERGADSNSIALQGLQKLPSTNNKFTYNCDNHTFNYLIDNGYTYC 182
Query: 67 VVAVESVGRQIPIAFLERIKDDFNKRYGGGRATT 100
VVA E+ GRQ+P+AFLER+KDDF +YGG +A T
Sbjct: 183 VVADETTGRQVPMAFLERVKDDFVSKYGGEKAPT 284
>BG585788 homologue to GP|7268326|emb SYBL1 like protein {Arabidopsis
thaliana}, partial (65%)
Length = 847
Score = 104 bits (260), Expect = 2e-23
Identities = 46/67 (68%), Positives = 58/67 (85%)
Frame = +2
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M Q+S IYSFVARGT++LAEY++FTGNF IA QCLQKLP+SNN+FTY+CD HTF+FLV+
Sbjct: 44 MSQESFIYSFVARGTMVLAEYTEFTGNFPAIAAQCLQKLPSSNNKFTYSCDHHTFNFLVE 223
Query: 61 NGFTYCV 67
+G+ V
Sbjct: 224 DGYVSLV 244
Score = 47.4 bits (111), Expect = 4e-06
Identities = 22/29 (75%), Positives = 24/29 (81%)
Frame = +1
Query: 66 CVVAVESVGRQIPIAFLERIKDDFNKRYG 94
CVVA ESV +QI IAFLER+K DF KRYG
Sbjct: 724 CVVAKESVSKQISIAFLERVKADFKKRYG 810
>BG648270 similar to GP|22137292|gb At1g04740/T1G11_1 {Arabidopsis thaliana},
partial (59%)
Length = 835
Score = 76.3 bits (186), Expect = 9e-15
Identities = 38/58 (65%), Positives = 44/58 (75%)
Frame = +1
Query: 65 YCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
YCVV VES R I +AFL+RIK+DF KRYGGG+A TAT+KSLNKEFG Q+CV
Sbjct: 358 YCVVGVESFDRHIAMAFLDRIKEDFTKRYGGGKAATATSKSLNKEFG--YSSL*QFCV 525
Score = 75.9 bits (185), Expect = 1e-14
Identities = 36/43 (83%), Positives = 40/43 (92%)
Frame = +2
Query: 110 FGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKV 152
F PKLKE MQYCVEHPEEVSKLAKVKAQVS+V+ VM+ENID+V
Sbjct: 593 FRPKLKEHMQYCVEHPEEVSKLAKVKAQVSQVQDVMLENIDQV 721
Score = 74.3 bits (181), Expect = 3e-14
Identities = 31/47 (65%), Positives = 41/47 (86%)
Frame = +1
Query: 18 LAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFT 64
LAE+++FTGNF IALQCLQ+LPA+N +FTYN DGHTF++L +GF+
Sbjct: 16 LAEHTNFTGNFVEIALQCLQRLPATNTKFTYNTDGHTFNYLAHDGFS 156
>BQ255326 similar to GP|9757817|dbj synaptobrevin-like protein {Arabidopsis
thaliana}, partial (75%)
Length = 543
Score = 73.6 bits (179), Expect = 6e-14
Identities = 46/121 (38%), Positives = 70/121 (57%), Gaps = 1/121 (0%)
Frame = +1
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA-SNNRFTYNCDGHTFSFLV 59
+ +++Y+ VARGTV+LAE+S TGN +A + L+KLP SN+R D + L
Sbjct: 7 LSSMAILYASVARGTVVLAEFSAGTGNTGAVARRLLEKLPTESNSRLCCXQDRYISHILR 186
Query: 60 DNGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQ 119
+G T +A ++ GR+IP + LE I+ F K Y A A A ++N EF L +QM+
Sbjct: 187 SDGLTCLRMANDTFGRRIPFSCLEDIQMRFMKNY-SRVANYAPAYAMNDEFSRVLHQQME 363
Query: 120 Y 120
+
Sbjct: 364 F 366
>TC87130 weakly similar to PIR|T04067|T04067 hypothetical protein F28M11.90
- Arabidopsis thaliana, partial (71%)
Length = 1510
Score = 54.3 bits (129), Expect = 4e-08
Identities = 25/89 (28%), Positives = 46/89 (51%)
Frame = +1
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNG 62
Q + Y V++G+ IL YS + CL+ +P+ + + T+ FLV++G
Sbjct: 400 QNKVHYCCVSKGSHILYAYSGGDKEVENVTALCLEAVPSFHRWYFETIGKRTYGFLVEDG 579
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNK 91
+ Y + E +G + + FLE ++D+F K
Sbjct: 580 YVYFTIVDEGLGNLVVLRFLEHVRDEFRK 666
Score = 38.5 bits (88), Expect = 0.002
Identities = 26/97 (26%), Positives = 45/97 (45%), Gaps = 10/97 (10%)
Frame = +3
Query: 133 KVKAQVSEVKGVMMENIDKVIDRGEKIEVL---------VDKTENLRSQAQDFR-QQGTQ 182
KVK V ++ V +E K DRG + + + +L+ R + G Q
Sbjct: 927 KVKDHVIAMRDVELEEHKKSTDRGSRPDSGNLDGISQGGAATSSSLQKDMGSMRTRSGPQ 1106
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
RK W++ ++I L + A + ++ +I L +C G SC
Sbjct: 1107 NVRKKWWRQVRIVLAIDAAVCIILFVIWLVICRGISC 1217
>AJ501851 similar to PIR|F86180|F861 hypothetical protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 121
Score = 48.1 bits (113), Expect = 3e-06
Identities = 23/28 (82%), Positives = 26/28 (92%)
Frame = +2
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNF 28
M QQSLIYSFVARG VILAE+++FTGNF
Sbjct: 35 MVQQSLIYSFVARGMVILAEHTNFTGNF 118
>BI267523
Length = 602
Score = 42.0 bits (97), Expect = 2e-04
Identities = 26/96 (27%), Positives = 49/96 (50%), Gaps = 10/96 (10%)
Frame = +2
Query: 1 MGQQSLI-------YSFVARGTVILAEYSDFT---GNFTTIALQCLQKLPASNNRFTYNC 50
MG S+I Y +A+ T ILA + + N T+A +CL++ P +++ F++
Sbjct: 221 MGAASMIWNPDLIQYLCIAKSTTILAHHINKNPKDSNIETLASKCLEQSPPNHSFFSHTV 400
Query: 51 DGHTFSFLVDNGFTYCVVAVESVGRQIPIAFLERIK 86
+ T++FL++ F + + + I FL RI+
Sbjct: 401 NNRTYTFLIEPPFVLFAIFDTHLLKSHAITFLNRIR 508
>TC78059 similar to GP|4206787|gb|AAD11808.1| syntaxin-related protein
Nt-syr1 {Nicotiana tabacum}, partial (94%)
Length = 1595
Score = 31.6 bits (70), Expect = 0.25
Identities = 28/121 (23%), Positives = 57/121 (46%), Gaps = 3/121 (2%)
Frame = +2
Query: 96 GRATTATAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDR 155
G + T K++ ++ + + +Q E + V ++ + + E+ V M+ V +
Sbjct: 716 GESETFLQKAIQQQGRANIMDTIQEIQERHDTVKEIER---NLMELHQVFMDMSVLVQSQ 886
Query: 156 GEKIEVLVDKTENLRSQAQDFRQQGTQ---LRRKMWYQNMKIKLIVLAIIIALILIIVLS 212
GE++ D E+ ++A + + G Q + RK K I + I++ +ILIIVL
Sbjct: 887 GEQL----DNIESHVARANSYVRGGVQQLHVARKHQMNTRKWTCIAIIILLIIILIIVLP 1054
Query: 213 V 213
+
Sbjct: 1055I 1057
>TC87204 small G-protein ROP9 [Medicago truncatula]
Length = 995
Score = 30.8 bits (68), Expect = 0.42
Identities = 18/71 (25%), Positives = 33/71 (46%), Gaps = 10/71 (14%)
Frame = +3
Query: 114 LKEQMQYCVEHP----------EEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLV 163
L++ Q+C++HP EE+ KL A + E EN+ V D ++ +
Sbjct: 516 LRDDKQFCIDHPGAVPITTAQGEELRKLINAPAYI-ECSSKSQENVKAVFDAAIRVVLQP 692
Query: 164 DKTENLRSQAQ 174
K + +++AQ
Sbjct: 693 PKQKKKKNKAQ 725
>BI265083 similar to GP|15450711|gb| At1g06210/F9P14_4 {Arabidopsis
thaliana}, partial (19%)
Length = 668
Score = 30.8 bits (68), Expect = 0.42
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 7/62 (11%)
Frame = +3
Query: 4 QSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQ-------KLPASNNRFTYNCDGHTFS 56
QS +Y F +G + + FT + ++CL K+P+S +R T +C G S
Sbjct: 354 QSDLYEFERKG*IT*HDDRKFTTYSVCLRVKCLSTSFRSP*KIPSSRSRATCSCSGRLGS 533
Query: 57 FL 58
FL
Sbjct: 534 FL 539
>TC87986 similar to GP|5669636|gb|AAD46403.1| ethylene-responsive elongation
factor EF-Ts precursor {Lycopersicon esculentum},
partial (45%)
Length = 897
Score = 30.0 bits (66), Expect = 0.72
Identities = 14/40 (35%), Positives = 26/40 (65%)
Frame = +2
Query: 110 FGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENI 149
FGP+ E+M+ +EHP +++ V+ ++EV +M EN+
Sbjct: 599 FGPQSLEEMKLNLEHP-KINGETTVQNAITEVAAIMGENV 715
>CA920901 homologue to GP|22597174|gb syntaxin {Glycine max}, partial (36%)
Length = 793
Score = 29.3 bits (64), Expect = 1.2
Identities = 23/90 (25%), Positives = 43/90 (47%), Gaps = 1/90 (1%)
Frame = -2
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQ-AQDFRQQGTQ 182
H E + +++ Q+ EV + + V ++G I+ + EN + AQ Q
Sbjct: 759 HWEREQGIQEIQQQIGEVNEIFKDLAVLVHEQGTMIDDIGSNIENSHAATAQAKSQLAKA 580
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLS 212
+ + ++ L+V+ I+ LILIIVL+
Sbjct: 579 SKTQRSNSSLACLLLVIFGIVLLILIIVLA 490
>TC80122 similar to GP|19401700|gb|AAL87667.1 transcription factor RAU1
{Oryza sativa}, partial (72%)
Length = 1127
Score = 28.1 bits (61), Expect = 2.7
Identities = 14/57 (24%), Positives = 32/57 (55%), Gaps = 3/57 (5%)
Frame = +2
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMME---NIDKVIDRGEKIEVLVDKTENLRSQAQ 174
C HP +++ + + ++SE + + N+DK + + +++ VD ++L+ QAQ
Sbjct: 392 CATHPRSIAERVR-RTKISERMRKLQDLVPNMDKQTNTADMLDLAVDYIKDLQKQAQ 559
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,760,186
Number of Sequences: 36976
Number of extensions: 65919
Number of successful extensions: 437
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 428
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 432
length of query: 219
length of database: 9,014,727
effective HSP length: 92
effective length of query: 127
effective length of database: 5,612,935
effective search space: 712842745
effective search space used: 712842745
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126019.3


