

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.13 - phase: 0
(367 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
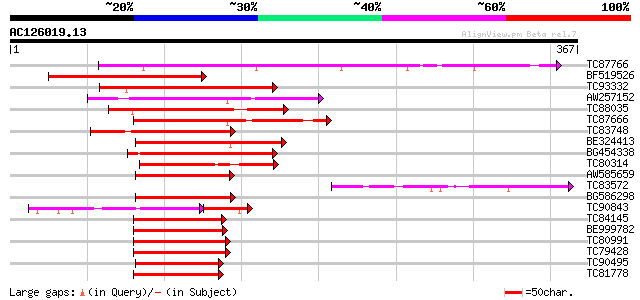
Score E
Sequences producing significant alignments: (bits) Value
TC87766 similar to GP|10177472|dbj|BAB10863. contains similarity... 193 1e-49
BF519526 similar to PIR|T02525|T025 probable DOF zinc finger pro... 152 2e-37
TC93332 similar to PIR|T48267|T48267 probable zinc finger protei... 147 8e-36
AW257152 similar to PIR|D86409|D864 hypothetical protein AAF9842... 134 6e-32
TC88035 similar to GP|6092016|dbj|BAA85655.1 elicitor-responsive... 133 1e-31
TC87666 homologue to GP|4996640|dbj|BAA78572.1 Dof zinc finger p... 125 2e-29
TC83748 homologue to PIR|T02375|T02375 finger protein BBF3 - com... 125 3e-29
BE324413 homologue to GP|10176912|dbj zinc finger protein {Arabi... 124 8e-29
BG454338 homologue to PIR|T52044|T520 dof zinc finger protein [i... 120 6e-28
TC80314 similar to PIR|D86409|D86409 hypothetical protein AAF984... 111 5e-25
AW585659 similar to GP|2262110|gb|A zinc finger protein isolog {... 110 7e-25
TC83572 similar to GP|20268789|gb|AAM14097.1 unknown protein {Ar... 109 2e-24
BG586298 homologue to PIR|T08455|T08 hypothetical protein F22O6.... 108 3e-24
TC90843 similar to GP|6092016|dbj|BAA85655.1 elicitor-responsive... 98 4e-24
TC84145 similar to GP|7242908|dbj|BAA92506.1 ESTs C23582(S11122)... 104 5e-23
BE999782 similar to PIR|T09661|T096 ascorbate oxidase promoter-b... 103 1e-22
TC80991 similar to GP|15983797|gb|AAL10495.1 AT5g39660/MIJ24_130... 102 2e-22
TC79428 similar to PIR|T09661|T09661 ascorbate oxidase promoter-... 102 3e-22
TC90495 similar to PIR|H84752|H84752 probable DOF zinc finger pr... 99 2e-21
TC81778 similar to GP|7242908|dbj|BAA92506.1 ESTs C23582(S11122)... 98 6e-21
>TC87766 similar to GP|10177472|dbj|BAB10863. contains similarity to dof
zinc finger protein~gene_id:MQB2.26 {Arabidopsis
thaliana}, partial (21%)
Length = 1516
Score = 193 bits (490), Expect = 1e-49
Identities = 125/326 (38%), Positives = 176/326 (53%), Gaps = 26/326 (7%)
Frame = +2
Query: 58 GGSTGSIRPGSMSDRARMANMPMPETAL--KCPRCESTNTKFCYFNNYSLSQPRHFCKTC 115
G GS RP SM+DRA+M+ + + A KCPRCESTNTKFCY+NNY+LSQPRHFCKTC
Sbjct: 239 GSYQGSRRPVSMTDRAKMSKIHQNDAAAAQKCPRCESTNTKFCYYNNYNLSQPRHFCKTC 418
Query: 116 RRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDR--------QTGSASSTN 167
RRYWT+GGALRNVPVGGG RRN + K ++ S P +D GS+S++
Sbjct: 419 RRYWTKGGALRNVPVGGGCRRNNKRRKGNSVSKSPLKPNHNDHLQIGTGAAAGGSSSASA 598
Query: 168 SITSNNNIHSSADILGLTPQMSSLRFMTPLHHNFSNPNDFVGGGDL---------GLNYG 218
+ +SNN +S +G+ + F+T LH + N G G L N
Sbjct: 599 NSSSNNGCTNSNVNIGMPNFPTQFPFVTSLHRHNDNSYASEGIGSLLAKNMSNSTTTNVE 778
Query: 219 FSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSM-THQQ---HQFPFLANLEGTNSNLYP 274
F G +G+ GG+ ++S+ EQWR P S+ HQQ QFPFL+NL+ L+
Sbjct: 779 FQLGSGSSSIGNGNGGSLLISNGIGEQWRFPNSLQIHQQQQYQQFPFLSNLQ-PQIGLFQ 955
Query: 275 FEGNVQNHEMITSGGYVRPKVISTS---GIMTQLASVKMEDSINRGFLGINTSNNNSNQG 331
F G +N+ + + S+S G+M Q+ +VK+E++ ++G + S
Sbjct: 956 FGG--ENNGEPQRNLFRSKETYSSSVSIGMMPQVNTVKVEENNHQGLSLPKNLLSGSGNS 1129
Query: 332 SEQFWNSAVVTGNSAANWTDHVSGFS 357
++ FWN GN N + V F+
Sbjct: 1130NDLFWN-----GNGNENAWNEVPSFT 1192
>BF519526 similar to PIR|T02525|T025 probable DOF zinc finger protein
[imported] - Arabidopsis thaliana, partial (26%)
Length = 453
Score = 152 bits (385), Expect = 2e-37
Identities = 69/102 (67%), Positives = 77/102 (74%)
Frame = +1
Query: 26 HHQQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMPMPETAL 85
HHQQP + + Q P P Q GGG+ GSIRPGSM+DRAR+A +P PE AL
Sbjct: 148 HHQQPNHQQANGSDNTQLLPPLPPQVGGSTTGGGTMGSIRPGSMADRARLAKLPPPEPAL 327
Query: 86 KCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRN 127
KCPRC+STNTKFCYFNNYSL+QPRHFCKTC RYWTRGGALRN
Sbjct: 328 KCPRCDSTNTKFCYFNNYSLTQPRHFCKTCMRYWTRGGALRN 453
>TC93332 similar to PIR|T48267|T48267 probable zinc finger protein -
Arabidopsis thaliana, partial (20%)
Length = 666
Score = 147 bits (370), Expect = 8e-36
Identities = 72/131 (54%), Positives = 91/131 (68%), Gaps = 16/131 (12%)
Frame = +1
Query: 59 GSTGSIRPGSMSDRAR---------------MANMPMP-ETALKCPRCESTNTKFCYFNN 102
G+ IRPGS+ D+A+ A +P P ET LKCPRCESTNTKFCY+NN
Sbjct: 1 GAGSIIRPGSIPDQAQAQAAHEVHTGQVQTHQAKLPPPSETNLKCPRCESTNTKFCYYNN 180
Query: 103 YSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGS 162
Y+LSQPRHFCKTCRRYWTRGGALRNVPVGGG RR+K++ KS+ + +KSP S+
Sbjct: 181 YNLSQPRHFCKTCRRYWTRGGALRNVPVGGGCRRSKKTKKSNTSSTPSKSPISNSNDKEY 360
Query: 163 ASSTNSITSNN 173
++S ++I S +
Sbjct: 361 STSASAIHSTS 393
>AW257152 similar to PIR|D86409|D864 hypothetical protein AAF98424.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 587
Score = 134 bits (337), Expect = 6e-32
Identities = 72/157 (45%), Positives = 92/157 (57%), Gaps = 4/157 (2%)
Frame = +1
Query: 51 SQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRH 110
S + GGG+ +PG + + AL+CPRC+S+NTKFCY+NNYSLSQPRH
Sbjct: 40 SMASSSGGGNRVMEKPGQEL-------LQQQQQALRCPRCDSSNTKFCYYNNYSLSQPRH 198
Query: 111 FCKTCRRYWTRGGALRNVPVGGGFRRNKR----SSKSSNNGNSTKSPASSDRQTGSASST 166
FCK C+RYWTRGG LRNVPVGGG R+NKR + SSN +T SP+SS SA+S
Sbjct: 199 FCKACKRYWTRGGTLRNVPVGGGCRKNKRLKRPNHSSSNIDTATASPSSS--TPNSANSN 372
Query: 167 NSITSNNNIHSSADILGLTPQMSSLRFMTPLHHNFSN 203
H S DI + ++SL + H+ N
Sbjct: 373 PPSQQQQQQHHSFDIASTSNHINSLLYGNSSCHDVMN 483
>TC88035 similar to GP|6092016|dbj|BAA85655.1 elicitor-responsive Dof
protein ERDP {Pisum sativum}, partial (77%)
Length = 1397
Score = 133 bits (334), Expect = 1e-31
Identities = 69/129 (53%), Positives = 81/129 (62%), Gaps = 13/129 (10%)
Frame = +3
Query: 65 RPGSMSDRARMANM-------------PMPETALKCPRCESTNTKFCYFNNYSLSQPRHF 111
+P MS ++ M NM P PE AL CPRC STNTKFCY+NNYSL+QPR+F
Sbjct: 138 QPQQMSSQSSMENMLGCSKEEQERKPKPQPEQALNCPRCNSTNTKFCYYNNYSLTQPRYF 317
Query: 112 CKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITS 171
CK+CRRYWT+GG LRNVPVGGG R+NKRSS SS+ +SS R + N
Sbjct: 318 CKSCRRYWTKGGTLRNVPVGGGCRKNKRSSPSSS--------SSSKRVQDQTFAPNLNPF 473
Query: 172 NNNIHSSAD 180
NN HSS D
Sbjct: 474 NNLPHSSYD 500
>TC87666 homologue to GP|4996640|dbj|BAA78572.1 Dof zinc finger protein
{Oryza sativa}, partial (20%)
Length = 1828
Score = 125 bits (315), Expect = 2e-29
Identities = 62/130 (47%), Positives = 82/130 (62%), Gaps = 2/130 (1%)
Frame = +2
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKR- 139
P+ + CPRC STNTKFCY+NNYSL+QPR+FCKTCRRYWT GG+LRNVPVGGG R+NK+
Sbjct: 692 PQEQINCPRCNSTNTKFCYYNNYSLTQPRYFCKTCRRYWTEGGSLRNVPVGGGSRKNKKI 871
Query: 140 -SSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPLH 198
SS ++ N +S+K P D + ++ + N I D+ P M + H
Sbjct: 872 PSSLANPNNSSSKIP---DLNPPTLQVSHLSSQNPKIQGGQDLNLAFPSMEN------YH 1024
Query: 199 HNFSNPNDFV 208
HN P+ +V
Sbjct: 1025HNHGMPSSYV 1054
>TC83748 homologue to PIR|T02375|T02375 finger protein BBF3 - common tobacco
(fragment), partial (60%)
Length = 658
Score = 125 bits (314), Expect = 3e-29
Identities = 59/95 (62%), Positives = 71/95 (74%), Gaps = 1/95 (1%)
Frame = +3
Query: 53 SQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFC 112
S P G RP SM +R P + +LKCPRC+ST+TKFCY+NNYSLSQPR+FC
Sbjct: 288 SSPMSGDMLTCSRPSSMIERRLR---PPHDLSLKCPRCDSTHTKFCYYNNYSLSQPRYFC 458
Query: 113 KTCRRYWTRGGALRNVPVGGGFRRNKR-SSKSSNN 146
KTCRRYWT+GG LRN+PVGGG R+NK+ SSK SN+
Sbjct: 459 KTCRRYWTKGGTLRNIPVGGGCRKNKKVSSKKSND 563
>BE324413 homologue to GP|10176912|dbj zinc finger protein {Arabidopsis
thaliana}, partial (21%)
Length = 339
Score = 124 bits (310), Expect = 8e-29
Identities = 54/100 (54%), Positives = 77/100 (77%), Gaps = 2/100 (2%)
Frame = +2
Query: 82 ETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSS 141
+ +KCPRC+S NTKFCY+NNY+LSQPRHFCK CRRYWT+GG LRNVPVGGG R++KRSS
Sbjct: 14 QEVMKCPRCDSINTKFCYYNNYNLSQPRHFCKNCRRYWTKGGVLRNVPVGGGCRKSKRSS 193
Query: 142 --KSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSA 179
K S++ +ST P + + S+++S + ++++ ++A
Sbjct: 194 KPKKSSSSDSTVLPTTPPEAPENKSNSHSSSESSSLTAAA 313
>BG454338 homologue to PIR|T52044|T520 dof zinc finger protein [imported] -
Arabidopsis thaliana, partial (37%)
Length = 678
Score = 120 bits (302), Expect = 6e-28
Identities = 57/99 (57%), Positives = 73/99 (73%), Gaps = 2/99 (2%)
Frame = +1
Query: 77 NMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRR 136
N P E LKCPRCES NTKFCY+NNY+LSQPRHFCK C+RYWT+GGALRN+PVGGG R+
Sbjct: 190 NFPEQEQ-LKCPRCESNNTKFCYYNNYNLSQPRHFCKNCKRYWTKGGALRNIPVGGGTRK 366
Query: 137 -NKRSSKSSNNGN-STKSPASSDRQTGSASSTNSITSNN 173
KRSS S + S+ SP++S + +A + S++ +
Sbjct: 367 VTKRSSNSKRSTTPSSSSPSASTTSSSAAKVSASVSETD 483
>TC80314 similar to PIR|D86409|D86409 hypothetical protein AAF98424.1
[imported] - Arabidopsis thaliana, partial (19%)
Length = 986
Score = 111 bits (277), Expect = 5e-25
Identities = 52/90 (57%), Positives = 66/90 (72%)
Frame = +1
Query: 85 LKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSS 144
L CPRC+STNTKFCY+NNY+ SQPRHFC+ C+R+WT+GG LRNVPVGGG R+NKR K
Sbjct: 448 LNCPRCDSTNTKFCYYNNYNKSQPRHFCRACKRHWTKGGTLRNVPVGGG-RKNKRIKKP- 621
Query: 145 NNGNSTKSPASSDRQTGSASSTNSITSNNN 174
+T +S+ T S + T SIT+ N+
Sbjct: 622 ----TTPVTSSTTITTTSTTCTTSITNMNS 699
>AW585659 similar to GP|2262110|gb|A zinc finger protein isolog {Arabidopsis
thaliana}, partial (27%)
Length = 483
Score = 110 bits (276), Expect = 7e-25
Identities = 43/64 (67%), Positives = 56/64 (87%)
Frame = +2
Query: 82 ETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSS 141
+ A+ CPRC S NTKFCY+NNYSL+QPR+FCKTCRRYWT+GG++RN+PVGGG R+N ++
Sbjct: 194 QEAINCPRCNSINTKFCYYNNYSLTQPRYFCKTCRRYWTQGGSIRNIPVGGGSRKNNKNR 373
Query: 142 KSSN 145
SS+
Sbjct: 374 SSSS 385
>TC83572 similar to GP|20268789|gb|AAM14097.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 819
Score = 109 bits (272), Expect = 2e-24
Identities = 73/169 (43%), Positives = 98/169 (57%), Gaps = 12/169 (7%)
Frame = -1
Query: 209 GGGDLGLNYGFSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSMTHQQHQFPFLANLEGT 268
G GDL + G S GGVG+ G G SIL S EQWRMP Q HQFPFL+ LE +
Sbjct: 804 GVGDLNFHIGSSLGGGVGNGG---GSASILPVSNFEQWRMP-----QTHQFPFLSTLEAS 649
Query: 269 NSN--LYPFEG-------NVQNHEMITSGGYVRPKVISTSGIMTQLASVKMEDSINRGFL 319
+S+ LYPFEG N Q H GG S +++ +SVKME++ +R FL
Sbjct: 648 SSHGLLYPFEGGGNGGGANDQGHAY---GG--------VSKVLSNNSSVKMEENQSRQFL 502
Query: 320 GI---NTSNNNSNQGSEQFWNSAVVTGNSAANWTDHVSGFSSSSNTTSN 365
G+ N +NNN+N SEQFW+S+ G + + WTD + ++SSS TT++
Sbjct: 501 GMINNNNNNNNNNPNSEQFWSSSAGGGANNSPWTDLSTAYNSSSATTNS 355
>BG586298 homologue to PIR|T08455|T08 hypothetical protein F22O6.180 -
Arabidopsis thaliana, partial (65%)
Length = 723
Score = 108 bits (271), Expect = 3e-24
Identities = 45/65 (69%), Positives = 54/65 (82%)
Frame = +3
Query: 82 ETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSS 141
E CPRC S+NTKFCY+NNYSL+QPR+FCK CRRYWT+GG+LRNVPVGGG R+++R
Sbjct: 180 EITPSCPRCGSSNTKFCYYNNYSLTQPRYFCKGCRRYWTKGGSLRNVPVGGGCRKSRRPK 359
Query: 142 KSSNN 146
SS N
Sbjct: 360 SSSGN 374
>TC90843 similar to GP|6092016|dbj|BAA85655.1 elicitor-responsive Dof
protein ERDP {Pisum sativum}, partial (19%)
Length = 698
Score = 97.8 bits (242), Expect(2) = 4e-24
Identities = 56/137 (40%), Positives = 73/137 (52%), Gaps = 23/137 (16%)
Frame = +1
Query: 13 PFCL------CCLLLQQQNHHQQP----GNGGSVSQQ-------------LLQTQTPPPQ 49
PFC CC L QQN + GNG S + LLQ T PP
Sbjct: 52 PFCFSVFFLDCCSLDPQQN*RKMIQELFGNGASATLTSSERNYSSINGGVLLQPTTQPPS 231
Query: 50 QSQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPR 109
S +T + P +++ ++ L+CPRC+S+NTKFCY+NNY+L+QPR
Sbjct: 232 SCPSST----TTTTTAPTTVTTTTTTSSENQQH--LRCPRCDSSNTKFCYYNNYNLTQPR 393
Query: 110 HFCKTCRRYWTRGGALR 126
HFCKTCRRYWT+GGAL+
Sbjct: 394 HFCKTCRRYWTKGGALK 444
Score = 31.2 bits (69), Expect(2) = 4e-24
Identities = 16/36 (44%), Positives = 22/36 (60%), Gaps = 4/36 (11%)
Frame = +2
Query: 126 RNVPVGGGFRRNKRSSKSSNNG----NSTKSPASSD 157
RNVP+GGG R+NK S+N N K+ +SS+
Sbjct: 443 RNVPIGGGCRKNKSIGASNNAAKIATNKMKTVSSSE 550
>TC84145 similar to GP|7242908|dbj|BAA92506.1 ESTs C23582(S11122)
AU056531(S20663) correspond to a region of the predicted
gene.~Similar to AOBP, partial (16%)
Length = 673
Score = 104 bits (260), Expect = 5e-23
Identities = 41/60 (68%), Positives = 49/60 (81%)
Frame = +3
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ L CPRC S TKFCYFNNY+++QPRHFCK C+RYWT GG +RNVP+G G RRNK+S
Sbjct: 231 PDKVLPCPRCNSLETKFCYFNNYNVNQPRHFCKNCQRYWTAGGVIRNVPIGAGKRRNKQS 410
>BE999782 similar to PIR|T09661|T096 ascorbate oxidase promoter-binding
protein AOBP - winter squash, partial (17%)
Length = 630
Score = 103 bits (256), Expect = 1e-22
Identities = 41/61 (67%), Positives = 51/61 (83%)
Frame = +2
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ L CPRC S +TKFCY+NNY+++QPRHFCK C+RYWT GGA+RNVPVG G R+NK S
Sbjct: 2 PDKILPCPRCYSMDTKFCYYNNYNVNQPRHFCKKCQRYWTAGGAMRNVPVGAGRRKNKNS 181
Query: 141 S 141
+
Sbjct: 182 A 184
>TC80991 similar to GP|15983797|gb|AAL10495.1 AT5g39660/MIJ24_130
{Arabidopsis thaliana}, partial (19%)
Length = 782
Score = 102 bits (255), Expect = 2e-22
Identities = 41/63 (65%), Positives = 50/63 (79%)
Frame = +3
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ + CPRC S +TKFCYFNNY+++QPRHFCK C+RYWT GG +RNVPVG G R+NK
Sbjct: 486 PDKIIPCPRCNSMDTKFCYFNNYNVNQPRHFCKKCQRYWTAGGTMRNVPVGAGRRKNKNL 665
Query: 141 SKS 143
S S
Sbjct: 666 STS 674
>TC79428 similar to PIR|T09661|T09661 ascorbate oxidase promoter-binding
protein AOBP - winter squash, partial (27%)
Length = 997
Score = 102 bits (253), Expect = 3e-22
Identities = 40/63 (63%), Positives = 51/63 (80%)
Frame = +1
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ L CPRC S +TKFCY+NNY+++QPR+FCK C+RYWT GG +RNVPVG G R+NK +
Sbjct: 511 PDKLLLCPRCNSADTKFCYYNNYNVNQPRYFCKACQRYWTAGGTMRNVPVGAGRRKNKNN 690
Query: 141 SKS 143
S S
Sbjct: 691 SSS 699
>TC90495 similar to PIR|H84752|H84752 probable DOF zinc finger protein
[imported] - Arabidopsis thaliana, partial (41%)
Length = 620
Score = 99.4 bits (246), Expect = 2e-21
Identities = 40/57 (70%), Positives = 48/57 (84%)
Frame = +3
Query: 82 ETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNK 138
E + CPRC+S TKFCYFNNY+++QPRHFCK+C+RYWT GGALRNVPVG G R+ K
Sbjct: 243 EKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKSCQRYWTAGGALRNVPVGAGRRKAK 413
>TC81778 similar to GP|7242908|dbj|BAA92506.1 ESTs C23582(S11122)
AU056531(S20663) correspond to a region of the predicted
gene.~Similar to AOBP, partial (17%)
Length = 1081
Score = 97.8 bits (242), Expect = 6e-21
Identities = 37/58 (63%), Positives = 49/58 (83%)
Frame = +3
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNK 138
P+ ++CPRC+S +TKFCYFNNY+++QPRHFC++C RYWT GG +RNV VG G R+NK
Sbjct: 291 PDKIVQCPRCKSFDTKFCYFNNYNVNQPRHFCRSCHRYWTNGGTMRNVAVGAGRRKNK 464
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.128 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,697,864
Number of Sequences: 36976
Number of extensions: 222223
Number of successful extensions: 1577
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 1521
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1561
length of query: 367
length of database: 9,014,727
effective HSP length: 97
effective length of query: 270
effective length of database: 5,428,055
effective search space: 1465574850
effective search space used: 1465574850
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC126019.13


