

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.10 + phase: 0
(435 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
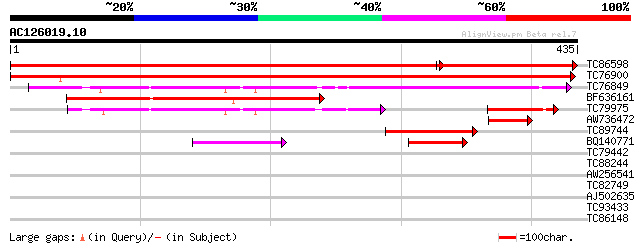
Score E
Sequences producing significant alignments: (bits) Value
TC86598 similar to GP|1129145|emb|CAA53078.1 acetyl-CoA C-acyltr... 638 0.0
TC76900 similar to GP|393707|emb|CAA47926.1| acetyl-CoA acyltra... 664 0.0
TC76849 similar to GP|19880269|gb|AAM00280.1 acetoacetyl-CoA thi... 218 5e-57
BF636161 weakly similar to SP|Q05493|THIK 3-ketoacyl-CoA thiolas... 180 1e-45
TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetylt... 101 5e-22
AW736472 homologue to SP|P21653|TIP1 Tonoplast intrinsic protein... 64 9e-11
TC89744 similar to GP|19880269|gb|AAM00280.1 acetoacetyl-CoA thi... 64 1e-10
BQ140771 similar to GP|13447074|gb| acetyl-CoA acetyl transferas... 49 5e-06
TC79442 homologue to GP|7385203|gb|AAF61731.1| beta-ketoacyl-ACP... 35 0.074
TC88244 homologue to GP|7385201|gb|AAF61730.1| beta-ketoacyl-ACP... 33 0.22
AW256541 30 1.8
TC82749 weakly similar to PIR|E70313|E70313 histidine triad-like... 30 1.8
AJ502635 weakly similar to PIR|E70313|E703 histidine triad-like ... 30 1.8
TC93433 similar to GP|11994337|dbj|BAB02296. lysophospholipase-l... 29 4.1
TC86148 homologue to PIR|T05747|T05747 histone H2A.M4I22.40 - Ar... 28 9.0
>TC86598 similar to GP|1129145|emb|CAA53078.1 acetyl-CoA C-acyltransferase
{Mangifera indica}, partial (89%)
Length = 1534
Score = 638 bits (1646), Expect(2) = 0.0
Identities = 329/333 (98%), Positives = 330/333 (98%)
Frame = +1
Query: 1 MEKAVERQRVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSENDVVIVAAYRTAICKAK 60
MEKAVERQRVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSENDVVIVAAYRTAICKAK
Sbjct: 49 MEKAVERQRVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSENDVVIVAAYRTAICKAK 228
Query: 61 RGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAAFYAGFP 120
RGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAAFYAGFP
Sbjct: 229 RGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAAFYAGFP 408
Query: 121 DTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKTNPKVEIF 180
DTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKTNPKVEIF
Sbjct: 409 DTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKTNPKVEIF 588
Query: 181 AQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIPVSTKFV 240
AQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIPVSTKFV
Sbjct: 589 AQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIPVSTKFV 768
Query: 241 DPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAAAVLLMKR 300
DPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAAAVLLMKR
Sbjct: 769 DPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAAAVLLMKR 948
Query: 301 SVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAF 333
SVAVQKGLPILGIFRSFSAVGVDPAVMG +F
Sbjct: 949 SVAVQKGLPILGIFRSFSAVGVDPAVMGCWSSF 1047
Score = 220 bits (560), Expect(2) = 0.0
Identities = 108/108 (100%), Positives = 108/108 (100%)
Frame = +2
Query: 328 GVGPAFAIPAAVKSAGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGH 387
GVGPAFAIPAAVKSAGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGH
Sbjct: 1031 GVGPAFAIPAAVKSAGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGH 1210
Query: 388 PLGATGARCVATLLNEMKRRGKDCRFGVISMCIGSGMGAAAVFERGDF 435
PLGATGARCVATLLNEMKRRGKDCRFGVISMCIGSGMGAAAVFERGDF
Sbjct: 1211 PLGATGARCVATLLNEMKRRGKDCRFGVISMCIGSGMGAAAVFERGDF 1354
>TC76900 similar to GP|393707|emb|CAA47926.1| acetyl-CoA acyltransferase
{Cucumis sativus}, partial (96%)
Length = 1780
Score = 664 bits (1714), Expect = 0.0
Identities = 339/441 (76%), Positives = 387/441 (86%), Gaps = 7/441 (1%)
Frame = +3
Query: 1 MEKAVERQRVLLQHLNPNSVTSSHHS-THLSASLCSAG------QTGGSENDVVIVAAYR 53
M+KA+ RQ+VLLQHLNP+S +S+ S T L+AS C AG +T +DVVIVAAYR
Sbjct: 57 MDKAINRQKVLLQHLNPSSFSSNDSSSTSLNASACLAGDSAAYHRTAAFGDDVVIVAAYR 236
Query: 54 TAICKAKRGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMA 113
TA+CKAKRGGFKDT DD+LA VLKAVIEKTN+ PSEVGDIVVG+VLG GS+RA ECRMA
Sbjct: 237 TALCKAKRGGFKDTHADDILAPVLKAVIEKTNLNPSEVGDIVVGSVLGAGSQRASECRMA 416
Query: 114 AFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKT 173
AFYAGFP+TVP+RTVNRQCSSGLQAVADVAA I+AGFYDIGIGAGLE MT + ++
Sbjct: 417 AFYAGFPETVPVRTVNRQCSSGLQAVADVAAAIRAGFYDIGIGAGLESMTTNPMAWDGSV 596
Query: 174 NPKVEIFAQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEII 233
NP+V++F QA++CLL MGITSENVA R+GV+R+EQD+AAVESHRRAAAATA+G+FKDEII
Sbjct: 597 NPRVKMFEQAQNCLLPMGITSENVASRFGVSRKEQDEAAVESHRRAAAATAAGRFKDEII 776
Query: 234 PVSTKFVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAA 293
PVSTK VDPKTGEEK + +SVDDGIRPNA++ DL KLK FKKDGTTTAGN+SQVSDGA
Sbjct: 777 PVSTKIVDPKTGEEKSVTISVDDGIRPNASVSDLGKLKAVFKKDGTTTAGNSSQVSDGAG 956
Query: 294 AVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYE 353
AVLLMKRS+A+QKGLPILG+FRSF AVGVDPA+MGVGPA AIP AVK+AGLEL +IDL+E
Sbjct: 957 AVLLMKRSIAMQKGLPILGVFRSFVAVGVDPAIMGVGPAAAIPVAVKAAGLELDDIDLFE 1136
Query: 354 INEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLLNEMKRRGKDCRF 413
INEAFASQFVY KLGLD K+NVNGGAMA GHPLGATGARCVATLL+EMK+RGKDCR+
Sbjct: 1137INEAFASQFVYCRNKLGLDSQKINVNGGAMAIGHPLGATGARCVATLLHEMKKRGKDCRY 1316
Query: 414 GVISMCIGSGMGAAAVFERGD 434
GVISMCIG+GMGAAAVFERGD
Sbjct: 1317GVISMCIGTGMGAAAVFERGD 1379
>TC76849 similar to GP|19880269|gb|AAM00280.1 acetoacetyl-CoA thiolase
{Arabidopsis thaliana}, partial (97%)
Length = 1866
Score = 218 bits (554), Expect = 5e-57
Identities = 156/435 (35%), Positives = 233/435 (52%), Gaps = 18/435 (4%)
Frame = +1
Query: 15 LNPNSVTSSHHSTHLSASLCSAGQTGGSE-NDVVIVAAYRTAICKAKRGGFKDTL---PD 70
L+ ++ S HLS + +A + DV IV RT + G F TL P
Sbjct: 91 LSATTLKISRFIFHLSMAPAAASSDSSIQPRDVCIVGVARTPM-----GAFLGTLSSVPA 255
Query: 71 DLLASV-LKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAAFYAGFPDTVPLRTVN 129
L S+ ++A +++ NV+P+ V ++ G VL +A R AA AG +V TVN
Sbjct: 256 TKLGSIAIEAALKRANVDPAIVEEVFFGNVLSANLGQA-PARQAALGAGLSKSVVCTTVN 432
Query: 130 RQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQ------DTISGVRKTNPKVEIFAQA 183
+ C+SG++A A I+ G D+ + G+E M+ + G R + + +
Sbjct: 433 KVCASGMKATMLAAQSIQLGINDVVVAGGMENMSSVPKYLAEARKGSRLGHDSL-VDGML 609
Query: 184 RDCL------LSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIPVST 237
+D L + MG+ +E A + +TR++QD AV+S R +A +G F EI PV
Sbjct: 610 KDGLWDVYKDVGMGVCAELCADNHSITREDQDNYAVQSFERGISAQENGYFGWEITPVE- 786
Query: 238 KFVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDG-TTTAGNASQVSDGAAAVL 296
V G I V D+G+ + L KL+P+FK+ G + TAGNAS +SDGAAA++
Sbjct: 787 --VSGGRGRPSTI-VDKDEGLG-KFDAAKLRKLRPSFKETGGSVTAGNASSISDGAAALV 954
Query: 297 LMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYEINE 356
L+ A++ GL ++ ++ DP + PA AIP A+ +AGLE S ID YEINE
Sbjct: 955 LVSGEKALKLGLQVIAKITGYADAAQDPELFTTAPAIAIPKAIANAGLETSKIDFYEINE 1134
Query: 357 AFASQFVYSCKKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLLNEMKRRGKDCRFGVI 416
AFA + + K LGLD KVNV+GGA++ GHPLG +GAR + TLL +K+ K+ ++GV
Sbjct: 1135AFAVVALANQKLLGLDSAKVNVHGGAVSLGHPLGCSGARILVTLLGVLKQ--KNGKYGVG 1308
Query: 417 SMCIGSGMGAAAVFE 431
+C G G +A V E
Sbjct: 1309GVCNGGGGASALVVE 1353
>BF636161 weakly similar to SP|Q05493|THIK 3-ketoacyl-CoA thiolase
peroxisomal precursor (EC 2.3.1.16) (Beta-
ketothiolase), partial (40%)
Length = 684
Score = 180 bits (456), Expect = 1e-45
Identities = 98/200 (49%), Positives = 131/200 (65%), Gaps = 2/200 (1%)
Frame = +2
Query: 44 NDVVIVAAYRTAICKAKRGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPG 103
+DVVIV+A RT I KAK+GG + P+DLLA+V K VI ++ ++P + D+ VG VL PG
Sbjct: 77 DDVVIVSALRTPITKAKKGGLAEACPEDLLAAVFKGVISESKIDPKLIEDVAVGNVLPPG 256
Query: 104 SERAIECRMAAFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMT 163
+ RMAA Y+G P+T + T+NRQCSSGL +V +A I +G DIGIGAG+E MT
Sbjct: 257 GGATV-ARMAALYSGIPNTAAVNTLNRQCSSGLASVNQIAMEISSGQIDIGIGAGVESMT 433
Query: 164 QDTISGV--RKTNPKVEIFAQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAA 221
Q+ +GV + + V +A DCL MGITSENVA+ Y VTR +QD A +S++RA
Sbjct: 434 QNYGAGVMPERFSDAVMENEEAADCLAPMGITSENVAEEYXVTRAKQDAFAAQSYQRAFK 613
Query: 222 ATASGKFKDEIIPVSTKFVD 241
A + GKFK EI+ V D
Sbjct: 614 AQSEGKFKSEIVTVKYNDTD 673
>TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetyltransferase
{Hevea brasiliensis}, partial (73%)
Length = 1019
Score = 101 bits (252), Expect = 5e-22
Identities = 86/262 (32%), Positives = 128/262 (48%), Gaps = 18/262 (6%)
Frame = +2
Query: 45 DVVIVAAYRTAICKAKRGGFKDTLPD----DLLASVLKAVIEKTNVEPSEVGDIVVGTVL 100
DV IV RT + GGF +L L + +KA +++ NV+PS V ++ G VL
Sbjct: 86 DVCIVGVGRTPM-----GGFLGSLSSLSATHLGSLAIKAALKRANVDPSLVQEVFFGNVL 250
Query: 101 GPGSERAIECRMAAFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLE 160
+A R AA AG P +V T+N+ CSSG++A A I+ G D+ + G+E
Sbjct: 251 SANLGQA-PARQAALGAGIPTSVICTTINKVCSSGMKATMLAAQTIQLGSNDVVVVGGME 427
Query: 161 CMTQ------DTISGVRKTNPKVEIFAQARDCL------LSMGITSENVAQRYGVTRQEQ 208
M+ + G R + + I +D L MG+ +E A ++ +TR +Q
Sbjct: 428 SMSNAPKYIAEARKGSRLGHDTI-IDGMLKDGLWDVYNDFGMGVCAEMCADQHAITRDQQ 604
Query: 209 DQAAVESHRRAAAATASGKFKDEIIPVSTKFVDPKTGEEK-QIIVSVDDGIRPNANLVDL 267
D A++S R +A G F EI+P V+ +G K +V D+G+ + L
Sbjct: 605 DSYAIQSFERGISAQNGGHFAWEIVP-----VEIFSGRGKPSTLVDKDEGL-GKFDAAKL 766
Query: 268 AKLKPAFKK-DGTTTAGNASQV 288
KL P FKK GT TAGNAS +
Sbjct: 767 RKLGPNFKKVGGTVTAGNASSI 832
Score = 60.1 bits (144), Expect = 2e-09
Identities = 28/55 (50%), Positives = 41/55 (73%)
Frame = +1
Query: 367 KKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLLNEMKRRGKDCRFGVISMCIG 421
K LGLDP K+NV+GGA++ GHPLG +GAR + TLL ++ +G ++GV ++C G
Sbjct: 856 KLLGLDPEKLNVHGGAVSLGHPLGCSGARILXTLLGVLRHKGG--KYGVGAICNG 1014
>AW736472 homologue to SP|P21653|TIP1 Tonoplast intrinsic protein
root-specific RB7-5A (RT-TIP). [Common tobacco]
{Nicotiana tabacum}, partial (43%)
Length = 691
Score = 64.3 bits (155), Expect = 9e-11
Identities = 29/34 (85%), Positives = 31/34 (90%)
Frame = -2
Query: 368 KLGLDPTKVNVNGGAMAFGHPLGATGARCVATLL 401
KLGLD K+NVNGGAMA GHPLGATGARCVATL+
Sbjct: 684 KLGLDSQKINVNGGAMAIGHPLGATGARCVATLI 583
>TC89744 similar to GP|19880269|gb|AAM00280.1 acetoacetyl-CoA thiolase
{Arabidopsis thaliana}, partial (34%)
Length = 1362
Score = 63.5 bits (153), Expect = 1e-10
Identities = 32/71 (45%), Positives = 43/71 (60%)
Frame = +1
Query: 289 SDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSN 348
SDGAAA++L+ A + GL ++ + F+ P PA AIP A+ +AGLE S
Sbjct: 1066 SDGAAALVLVSEKKAHELGLHVIAKIKGFADAAXAPEFFTTAPALAIPKAISNAGLEASQ 1245
Query: 349 IDLYEINEAFA 359
ID YEINEAF+
Sbjct: 1246 IDYYEINEAFS 1278
>BQ140771 similar to GP|13447074|gb| acetyl-CoA acetyl transferase {Laccaria
bicolor}, partial (10%)
Length = 349
Score = 48.5 bits (114), Expect = 5e-06
Identities = 29/73 (39%), Positives = 44/73 (59%), Gaps = 1/73 (1%)
Frame = +1
Query: 141 DVAAYIKAGFYDIGIGAGLECM-TQDTISGVRKTNPKVEIFAQARDCLLSMGITSENVAQ 199
D+A IK+G +IGIGAG E M TQ + V + + +E +A +C + MG+ SE +A+
Sbjct: 130 DIANAIKSGMIEIGIGAGSESMSTQYGPNAVSELSELLENDKEAANCKVPMGVLSEAMAK 309
Query: 200 RYGVTRQEQDQAA 212
+TR +QD A
Sbjct: 310 DKXITRADQDAFA 348
Score = 44.3 bits (103), Expect = 9e-05
Identities = 21/45 (46%), Positives = 28/45 (61%)
Frame = +1
Query: 307 GLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDL 351
G I+G F S VGV P +MGVGPA AIP ++ GL ++D+
Sbjct: 1 GQKIIGKFVQASIVGVPPLLMGVGPAAAIPVVLEKTGLNKDDVDI 135
>TC79442 homologue to GP|7385203|gb|AAF61731.1| beta-ketoacyl-ACP synthetase
I-2 {Glycine max}, partial (41%)
Length = 1065
Score = 34.7 bits (78), Expect = 0.074
Identities = 31/126 (24%), Positives = 57/126 (44%), Gaps = 13/126 (10%)
Frame = +1
Query: 288 VSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVG------VDPAVMGVGPAFAIPAAVKS 341
+ +GA +++ A+++G PI+ + AV DP G+G + I +++
Sbjct: 37 MGEGAGVLVMESLEHAMKRGAPIIAEYLG-GAVNCDAYHMTDPRSDGLGVSSCIQSSLVD 213
Query: 342 AGLELSNIDLYEINEAFASQFV------YSCKKLGLDPTKVNVNGGAMAFGHPLGATGA- 394
AG+ ++ IN S V + KK+ +P+ + +N GH LGA G
Sbjct: 214 AGVSPEEVNY--INAHATSTLVGDLAEINAIKKVFKNPSGIKINATKSMIGHCLGAAGGL 387
Query: 395 RCVATL 400
+AT+
Sbjct: 388 EAIATI 405
>TC88244 homologue to GP|7385201|gb|AAF61730.1| beta-ketoacyl-ACP synthetase I
{Glycine max}, partial (89%)
Length = 1768
Score = 33.1 bits (74), Expect = 0.22
Identities = 30/126 (23%), Positives = 57/126 (44%), Gaps = 13/126 (10%)
Frame = +3
Query: 288 VSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVG------VDPAVMGVGPAFAIPAAVKS 341
+ +G+ ++L A+++G PI+ + AV DP G+G + I ++++
Sbjct: 912 MGEGSGVLVLESLEHAMKRGAPIIAEYLG-GAVNCDAYHMTDPRSDGLGVSTCIQSSLED 1088
Query: 342 AGLELSNIDLYEINEAFASQF------VYSCKKLGLDPTKVNVNGGAMAFGHPLGA-TGA 394
AG+ ++ IN S + + KK+ D + + +N GH LGA G
Sbjct: 1089 AGVSPEEVNY--INAHATSTLAGDLAEINAIKKVFKDTSGIKINATKSMIGHCLGAXXGL 1262
Query: 395 RCVATL 400
+AT+
Sbjct: 1263 EAIATV 1280
>AW256541
Length = 646
Score = 30.0 bits (66), Expect = 1.8
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Frame = +3
Query: 1 MEKAVERQ-RVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSEN 44
+E+A R+ VL HLNP T +S LSASLCSA N
Sbjct: 63 IEEATRRRVTVLSSHLNPTRFT---YSDPLSASLCSARSDSDDNN 188
>TC82749 weakly similar to PIR|E70313|E70313 histidine triad-like protein
pkcI - Aquifex aeolicus, partial (35%)
Length = 1018
Score = 30.0 bits (66), Expect = 1.8
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Frame = +1
Query: 1 MEKAVERQ-RVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSEN 44
+E+A R+ VL HLNP T +S LSASLCSA N
Sbjct: 106 IEEATRRRVTVLSSHLNPTRFT---YSDPLSASLCSARSDSDDNN 231
>AJ502635 weakly similar to PIR|E70313|E703 histidine triad-like protein pkcI
- Aquifex aeolicus, partial (35%)
Length = 460
Score = 30.0 bits (66), Expect = 1.8
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Frame = +3
Query: 1 MEKAVERQ-RVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSEN 44
+E+A R+ VL HLNP T +S LSASLCSA N
Sbjct: 93 IEEATRRRVTVLSSHLNPTRFT---YSDPLSASLCSARSDSDDNN 218
>TC93433 similar to GP|11994337|dbj|BAB02296. lysophospholipase-like protein
{Arabidopsis thaliana}, partial (69%)
Length = 753
Score = 28.9 bits (63), Expect = 4.1
Identities = 18/37 (48%), Positives = 21/37 (56%), Gaps = 5/37 (13%)
Frame = -2
Query: 8 QRVLLQHLNPNSVTSSHHSTHLSASL-----CSAGQT 39
Q VLL+HLNP +H HLS L CSAG+T
Sbjct: 479 QYVLLKHLNP-----PNHLDHLSGKLSHIKPCSAGKT 384
>TC86148 homologue to PIR|T05747|T05747 histone H2A.M4I22.40 - Arabidopsis
thaliana, partial (96%)
Length = 696
Score = 27.7 bits (60), Expect = 9.0
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = +2
Query: 122 TVPLRTVNRQCSSGLQ-AVADVAAYIKAGFYDIGIGAG 158
+ P + +R +GLQ V +A ++KAG Y +GAG
Sbjct: 119 SAPKKATSRSSKAGLQFPVGRIARFLKAGKYAERVGAG 232
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,283,972
Number of Sequences: 36976
Number of extensions: 116363
Number of successful extensions: 639
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 629
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 631
length of query: 435
length of database: 9,014,727
effective HSP length: 99
effective length of query: 336
effective length of database: 5,354,103
effective search space: 1798978608
effective search space used: 1798978608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126019.10


