

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
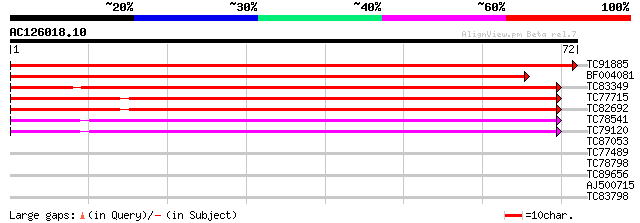
Query= AC126018.10 + phase: 0
(72 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91885 similar to SP|Q9BBQ9|CLPP_LOTJA ATP-dependent Clp protea... 144 8e-36
BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease pr... 133 1e-32
TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent prote... 55 4e-09
TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 pr... 50 2e-07
TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp protein... 50 2e-07
TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp p... 47 1e-06
TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp p... 44 2e-05
TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinas... 35 0.004
TC77489 MTD1 30 0.13
TC78798 similar to GP|21593878|gb|AAM65845.1 RALF precursor {Ara... 26 2.5
TC89656 25 4.3
AJ500715 25 7.4
TC83798 weakly similar to GP|10177750|dbj|BAB11063. cytochrome P... 25 7.4
>TC91885 similar to SP|Q9BBQ9|CLPP_LOTJA ATP-dependent Clp protease
proteolytic subunit (EC 3.4.21.92) (Endopeptidase Clp).
{Lotus japonicus}, partial (37%)
Length = 689
Score = 144 bits (362), Expect = 8e-36
Identities = 71/72 (98%), Positives = 72/72 (99%)
Frame = +2
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE
Sbjct: 467 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 646
Query: 61 AEAHGIVDTVAA 72
AEA+GIVDTVAA
Sbjct: 647 AEAYGIVDTVAA 682
>BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp). [Common
tobacco], partial (96%)
Length = 574
Score = 133 bits (335), Expect = 1e-32
Identities = 65/66 (98%), Positives = 66/66 (99%)
Frame = -2
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE
Sbjct: 198 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 19
Query: 61 AEAHGI 66
AEA+GI
Sbjct: 18 AEAYGI 1
>TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent protease
proteolytic subunit ClpP-like protein {Arabidopsis
thaliana}, partial (43%)
Length = 814
Score = 55.5 bits (132), Expect = 4e-09
Identities = 26/70 (37%), Positives = 44/70 (62%)
Frame = +1
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP+ Y GQ + + +++++M + +Y + TG+ I ++M+RD FM+AEE
Sbjct: 115 MIHQPSGG-YSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEE 291
Query: 61 AEAHGIVDTV 70
A+ GI+D V
Sbjct: 292 AKEFGIIDEV 321
>TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 proteolytic
subunit {Lycopersicon esculentum}, partial (76%)
Length = 1149
Score = 50.1 bits (118), Expect = 2e-07
Identities = 25/70 (35%), Positives = 43/70 (60%)
Frame = +3
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP GQ + ++A E+L + + + +TG++ +I +D +RD FMSA+E
Sbjct: 708 MIHQPLGGFQGGQT-DIDIQANEMLHHKANLNGYLSYQTGQSLKKINQDTDRDFFMSAKE 884
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 885 AKEYGLIDGV 914
>TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp proteinase (EC
3.4.21.-) chain P1 [imported] - Arabidopsis thaliana,
partial (73%)
Length = 1115
Score = 50.1 bits (118), Expect = 2e-07
Identities = 25/70 (35%), Positives = 43/70 (60%)
Frame = +2
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP GQ + ++A E+L + + + +TG++ +I +D +RD FMSA+E
Sbjct: 698 MIHQPLGGFQGGQT-DIDIQANEMLHHKANLNGYLSYQTGQSLEKINQDTDRDFFMSAKE 874
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 875 AKEYGLIDGV 904
>TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp protease;
nClpP3 {Arabidopsis thaliana}, partial (64%)
Length = 1158
Score = 47.4 bits (111), Expect = 1e-06
Identities = 28/70 (40%), Positives = 40/70 (57%)
Frame = +3
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP S G+ E + A EL+ + I I ++ TGK QI D +RD FM+ E
Sbjct: 591 MIHQPLGSA-GGKATEMSIRARELMYHKIKINKILSRITGKPEEQIELDTDRDNFMNPWE 767
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 768 AKEYGLIDDV 797
>TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp
protease-like protein {Arabidopsis thaliana}, partial
(67%)
Length = 1110
Score = 43.5 bits (101), Expect = 2e-05
Identities = 22/70 (31%), Positives = 42/70 (59%)
Frame = +1
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MI QP GQ + ++A+E++ + ++ I + TG++ Q+ +D++RD ++S E
Sbjct: 574 MIAQPPGGA-SGQAIDVEIQAKEIMHNKNNVSRIISSFTGRSLEQVLKDIDRDRYLSPIE 750
Query: 61 AEAHGIVDTV 70
A +GI+D V
Sbjct: 751 AVEYGIIDGV 780
>TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase (EC
3.4.21.-) catalytic chain P2 [imported] - Arabidopsis
thaliana, partial (74%)
Length = 1135
Score = 35.4 bits (80), Expect = 0.004
Identities = 21/70 (30%), Positives = 38/70 (54%)
Frame = +3
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
+I PA + G+ + EA ELL++R + A +TG+ +I D++R A+E
Sbjct: 678 VIQSPAGAA-RGRADDIQNEAAELLRIRDYLFTELANKTGQPVEKITEDLKRVKRFDAQE 854
Query: 61 AEAHGIVDTV 70
A +G++D +
Sbjct: 855 ALDYGLIDKI 884
>TC77489 MTD1
Length = 988
Score = 30.4 bits (67), Expect = 0.13
Identities = 14/39 (35%), Positives = 23/39 (58%)
Frame = +2
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRT 39
M A S Y G +CM EE+L +R++I+N Y+ ++
Sbjct: 263 MDENEAESKYNGGALDCMEALEEVLPIRRSISNFYSGKS 379
>TC78798 similar to GP|21593878|gb|AAM65845.1 RALF precursor {Arabidopsis
thaliana}, partial (65%)
Length = 885
Score = 26.2 bits (56), Expect = 2.5
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = +1
Query: 2 IHQPATSLYEGQVGECMLEAEE 23
IHQ T+ EG + +CML+ E
Sbjct: 220 IHQTKTATCEGSIADCMLQQGE 285
>TC89656
Length = 869
Score = 25.4 bits (54), Expect = 4.3
Identities = 12/40 (30%), Positives = 24/40 (60%)
Frame = +3
Query: 15 GECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDL 54
G C+ E EE+L T+ ++AQR S ++ + ++++L
Sbjct: 204 GSCLEEVEEVLL*AITLEGLHAQRKP*MSMRLKQKLKKNL 323
>AJ500715
Length = 576
Score = 24.6 bits (52), Expect = 7.4
Identities = 8/24 (33%), Positives = 15/24 (62%)
Frame = -1
Query: 24 LLKMRKTITNIYAQRTGKASWQIY 47
LLK +++ N+Y + + WQI+
Sbjct: 282 LLKRERSLMNVYLNQIMRTGWQIW 211
>TC83798 weakly similar to GP|10177750|dbj|BAB11063. cytochrome P450
{Arabidopsis thaliana}, partial (25%)
Length = 762
Score = 24.6 bits (52), Expect = 7.4
Identities = 13/61 (21%), Positives = 24/61 (39%)
Frame = +1
Query: 2 IHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEA 61
+H A +C + ++ +T+ N+YA SW + + F+ EE
Sbjct: 193 LHPTAPFALRQSSEDCKINGYDIKAHTRTLVNVYAIMRDPQSWVNPEEYIPERFLVGEEH 372
Query: 62 E 62
E
Sbjct: 373 E 375
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.129 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,670,464
Number of Sequences: 36976
Number of extensions: 12989
Number of successful extensions: 56
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56
length of query: 72
length of database: 9,014,727
effective HSP length: 48
effective length of query: 24
effective length of database: 7,239,879
effective search space: 173757096
effective search space used: 173757096
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC126018.10


