

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.10 + phase: 0 /pseudo
(498 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
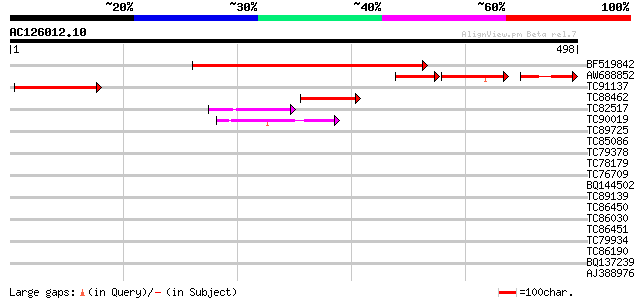
Score E
Sequences producing significant alignments: (bits) Value
BF519842 similar to GP|22136022|gb| DNA-binding protein-like {Ar... 409 e-114
AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding ... 89 9e-42
TC91137 similar to PIR|D71443|D71443 hypothetical protein - Arab... 158 5e-39
TC88462 67 1e-11
TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively ph... 49 3e-06
TC90019 weakly similar to GP|15010634|gb|AAK73976.1 At2g30580/T6... 42 5e-04
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 38 0.010
TC85086 similar to GP|4510376|gb|AAD21464.1| unknown protein {Ar... 36 0.030
TC79378 similar to PIR|T00518|T00518 hypothetical protein At2g23... 32 0.56
TC78179 weakly similar to GP|7211427|gb|AAF40306.1| RNA helicase... 32 0.56
TC76709 similar to GP|20259017|gb|AAM14224.1 unknown protein {Ar... 31 0.96
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 31 1.3
TC89139 similar to GP|14039799|gb|AAK53390.1 CLC-e chloride chan... 30 1.6
TC86450 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsi... 30 2.1
TC86030 similar to GP|15293277|gb|AAK93749.1 putative NADH-ubiqu... 30 2.1
TC86451 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsi... 30 2.1
TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 prote... 30 2.8
TC86190 weakly similar to PIR|T08454|T08454 hypothetical protein... 30 2.8
BQ137239 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 30 2.8
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 29 3.7
>BF519842 similar to GP|22136022|gb| DNA-binding protein-like {Arabidopsis
thaliana}, partial (19%)
Length = 623
Score = 409 bits (1051), Expect = e-114
Identities = 206/207 (99%), Positives = 206/207 (99%)
Frame = +3
Query: 161 DKEMEGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVC 220
DKEMEGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVC
Sbjct: 3 DKEMEGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVC 182
Query: 221 LATNILADDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSA 280
LATNILADDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSA
Sbjct: 183 LATNILADDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSA 362
Query: 281 TSKGEPKVSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPE 340
TSKGEPKVSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPE
Sbjct: 363 TSKGEPKVSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPE 542
Query: 341 PASQGSAKLVEEEVQQKLVPTDGGKKK 367
PASQGSAKLVEEEVQQKLVPTD GKKK
Sbjct: 543 PASQGSAKLVEEEVQQKLVPTDRGKKK 623
>AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding protein -
Arabidopsis thaliana, partial (9%)
Length = 536
Score = 89.0 bits (219), Expect(3) = 9e-42
Identities = 44/68 (64%), Positives = 47/68 (68%), Gaps = 9/68 (13%)
Frame = +1
Query: 380 L*FILEWHATLFGWIYGTICCSNDNDGLWSRPL*HAI---------CRWSASRPIWHAWL 430
L* ILEWHATL GWIYGT+C SN DGLWS PL HAI C+W+ASRPIWHA L
Sbjct: 187 L*SILEWHATLHGWIYGTVCWSNAYDGLWSWPLRHAISKWHAS*PVCKWNASRPIWHARL 366
Query: 431 RDACSSTS 438
DA S S
Sbjct: 367 HDASYSAS 390
Score = 68.2 bits (165), Expect(3) = 9e-42
Identities = 32/38 (84%), Positives = 35/38 (91%)
Frame = +2
Query: 340 EPASQGSAKLVEEEVQQKLVPTDGGKKKKKKKVRMPAN 377
EP SQGSA++VEEEVQQKLVPT+ GKKKKKKKVRMP N
Sbjct: 2 EPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTN 115
Score = 52.4 bits (124), Expect(3) = 9e-42
Identities = 29/51 (56%), Positives = 35/51 (67%), Gaps = 1/51 (1%)
Frame = +2
Query: 449 RDFAEFNIGMNVPPPVMNMNREPVMNKNREDFEARNA-ILKKQENERRVER 498
RD AEF++GMNVPPP M +RE+FEAR A +K+ENERRVER
Sbjct: 398 RDLAEFSMGMNVPPPAM----------SREEFEARKADARRKRENERRVER 520
>TC91137 similar to PIR|D71443|D71443 hypothetical protein - Arabidopsis
thaliana, partial (41%)
Length = 812
Score = 158 bits (399), Expect = 5e-39
Identities = 75/76 (98%), Positives = 76/76 (99%)
Frame = +3
Query: 5 ENLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSDF 64
+NLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSDF
Sbjct: 585 KNLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSDF 764
Query: 65 GAGRGFRRGAVGGRIG 80
GAGRGFRRGAVGGRIG
Sbjct: 765 GAGRGFRRGAVGGRIG 812
>TC88462
Length = 1157
Score = 67.4 bits (163), Expect = 1e-11
Identities = 35/53 (66%), Positives = 41/53 (77%)
Frame = -3
Query: 256 STYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANIQEIVAERKEV 308
S+ +++ M SS CPQ K+P PT SATSKGEPKVS VN+GM NIQEI ERK V
Sbjct: 735 SS*RIRYMESSCCPQSKLPFPTLSATSKGEPKVSLVNDGMMNIQEIADERKLV 577
>TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively
photomorphogenic 1 protein {Pisum sativum}, partial
(22%)
Length = 671
Score = 49.3 bits (116), Expect = 3e-06
Identities = 26/77 (33%), Positives = 39/77 (49%)
Frame = +3
Query: 175 DLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNK 234
+L + CP+C ++KDA LTS C SFC CI ++ +KS C C + +L PN
Sbjct: 138 ELDKDFLCPICMQIIKDAFLTS--CGHSFCYMCIITHLRNKSDCPCCGHYLTNSNLFPNF 311
Query: 235 TLRDAIHRILESGNSST 251
L + + + S T
Sbjct: 312 LLDKLLKKTSDRQISKT 362
>TC90019 weakly similar to GP|15010634|gb|AAK73976.1 At2g30580/T6B20.7
{Arabidopsis thaliana}, partial (32%)
Length = 1045
Score = 42.0 bits (97), Expect = 5e-04
Identities = 30/113 (26%), Positives = 48/113 (41%), Gaps = 5/113 (4%)
Frame = +2
Query: 182 CPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNI-----LADDLLPNKTL 236
CPLC+ ++KDA T C +FC KCI + + C NI + L P+ L
Sbjct: 233 CPLCHKLLKDAT-TISLCLHTFCRKCIHEKLSDEEVDSCPVCNIDLGILPVEKLTPDHNL 409
Query: 237 RDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVS 289
+D +I + +++++S PK + S+ PKVS
Sbjct: 410 QDIRTKIFPCKRQKV-------KAEEVVTSTPLPPKRKERSLSSLVVSAPKVS 547
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 37.7 bits (86), Expect = 0.010
Identities = 21/49 (42%), Positives = 23/49 (46%)
Frame = +1
Query: 65 GAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPT 113
G G G R G GG GGG G G C+ C SGHF + CPT
Sbjct: 67 GGGGGGRYGGGGGGGGGGGGGGS----------CYSCGESGHFARDCPT 183
>TC85086 similar to GP|4510376|gb|AAD21464.1| unknown protein {Arabidopsis
thaliana}, partial (15%)
Length = 656
Score = 36.2 bits (82), Expect = 0.030
Identities = 35/165 (21%), Positives = 70/165 (42%), Gaps = 17/165 (10%)
Frame = +3
Query: 175 DLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATN----ILADDL 230
++PP CP+ +M++ V T ++ K I ++M CVC TN ++ L
Sbjct: 126 EIPPYFVCPISFQIMEEPVTTVTGI--TYDRKSIEKWLMKAKICVCPVTNQSLPRSSEYL 299
Query: 231 LPNKTLRDAIH--------RILESGNSSTENAGSTYQVQDMLSS----RCPQPKIPSPTS 278
PN TL+ I +++++ S ++ + +Q ++ + C Q +
Sbjct: 300 TPNHTLQRLIKAWILSNEAKVVDNQIQSPKSPLNRIHLQKLVKNLELPNCFQASMEKILE 479
Query: 279 SATSKGEPKVSQVNEGMANIQEIVAERK-EVSATQQVSEQVKIPR 322
A ++ V G+ +V ++K + T + E +KI R
Sbjct: 480 LAKQSDRNRICMVEVGVTKAMVMVIKKKFKEXNTNGLXEALKIIR 614
>TC79378 similar to PIR|T00518|T00518 hypothetical protein At2g23140
[imported] - Arabidopsis thaliana, partial (36%)
Length = 2133
Score = 32.0 bits (71), Expect = 0.56
Identities = 22/82 (26%), Positives = 40/82 (47%), Gaps = 1/82 (1%)
Frame = +1
Query: 168 SSTRSVGDLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYI-MSKSACVCLATNIL 226
S + S +PP+ CPL +M D V+ + +++ I+N+I + + C +
Sbjct: 808 SQSNSPVPVPPDFCCPLSLELMTDPVIVAS--GQTYERAFIKNWIDLGLTVCPKTHQTLA 981
Query: 227 ADDLLPNKTLRDAIHRILESGN 248
+L+PN T++ I ES N
Sbjct: 982 HTNLIPNYTVKALIANWCESNN 1047
>TC78179 weakly similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna
radiata}, partial (12%)
Length = 692
Score = 32.0 bits (71), Expect = 0.56
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 11/52 (21%)
Frame = +2
Query: 46 IKALIDTPALDWQQQGSD--FGAGRGF---------RRGAVGGRIGGGRGFG 86
+K + P+L +++ S FG GRGF R G GG GGG+G+G
Sbjct: 173 LKVAKELPSLQQREESSGRRFGGGRGFSGGRGGGGYRFGGKGGGGGGGKGYG 328
>TC76709 similar to GP|20259017|gb|AAM14224.1 unknown protein {Arabidopsis
thaliana}, partial (87%)
Length = 2358
Score = 31.2 bits (69), Expect = 0.96
Identities = 15/47 (31%), Positives = 24/47 (50%)
Frame = +3
Query: 180 LHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNIL 226
L+C C+ K + TS CF S D + IMS + C+ +T ++
Sbjct: 2181 LYCSFCHGFTKPEITTSCICFLSKSDCTLLLRIMSSTFCISGSTFVI 2321
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 30.8 bits (68), Expect = 1.3
Identities = 20/43 (46%), Positives = 22/43 (50%)
Frame = +1
Query: 65 GAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHF 107
GAG G GA GGR+GGG G G R + P RC G F
Sbjct: 256 GAGGGPGPGAAGGRVGGG-GRGRGRGSGP------RCAALGEF 363
>TC89139 similar to GP|14039799|gb|AAK53390.1 CLC-e chloride channel protein
{Arabidopsis thaliana}, partial (24%)
Length = 1216
Score = 30.4 bits (67), Expect = 1.6
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = +3
Query: 395 YGTICCSNDNDGLWS 409
YGT+ CSND++G WS
Sbjct: 948 YGTLSCSNDHEGTWS 992
>TC86450 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsis
thaliana}, partial (39%)
Length = 1268
Score = 30.0 bits (66), Expect = 2.1
Identities = 15/54 (27%), Positives = 24/54 (43%)
Frame = +3
Query: 178 PELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLL 231
P CP+C M + + T C FC CI+ I +++ C I +L+
Sbjct: 729 PVFTCPICMGPMVEEMSTR--CGHIFCKSCIKAAISAQAKCPTCRKKITVKELI 884
>TC86030 similar to GP|15293277|gb|AAK93749.1 putative NADH-ubiquinone
oxireductase {Arabidopsis thaliana}, partial (88%)
Length = 1522
Score = 30.0 bits (66), Expect = 2.1
Identities = 18/57 (31%), Positives = 28/57 (48%), Gaps = 6/57 (10%)
Frame = +2
Query: 398 ICCSNDNDGLWS-RPL*H---AICRWSASRPIWHAWL--RDACSSTS*SNIMNMTVF 448
+C + G WS RP + IC+W IW W+ CS+T *+ I +++ F
Sbjct: 194 LCFHHSYQGRWSPRPQGYWWKIICQWYCCHSIWSNWIPWSLCCSTTC*NGISSLSSF 364
>TC86451 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsis
thaliana}, partial (51%)
Length = 995
Score = 30.0 bits (66), Expect = 2.1
Identities = 15/54 (27%), Positives = 24/54 (43%)
Frame = +2
Query: 178 PELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLL 231
P CP+C M + + T C FC CI+ I +++ C I +L+
Sbjct: 641 PVFTCPICMGPMVEEMSTR--CGHIFCKSCIKAAISAQAKCPTCRKKITVKELI 796
>TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 protein
{Arabidopsis thaliana}, partial (87%)
Length = 814
Score = 29.6 bits (65), Expect = 2.8
Identities = 22/62 (35%), Positives = 27/62 (43%), Gaps = 10/62 (16%)
Frame = +1
Query: 35 PLTSKADEDSKIKALIDTPALDWQQQGSDFGAGR--GFR--------RGAVGGRIGGGRG 84
PL+ + + I + P +G F GR GFR RG GGR GGGRG
Sbjct: 4 PLSKLSSQSHYILSQTMRPPARGGGRGGGFRGGRDGGFRGGRDGGGFRGRGGGRFGGGRG 183
Query: 85 FG 86
G
Sbjct: 184GG 189
>TC86190 weakly similar to PIR|T08454|T08454 hypothetical protein F22O6.170
- Arabidopsis thaliana, partial (60%)
Length = 1159
Score = 29.6 bits (65), Expect = 2.8
Identities = 23/100 (23%), Positives = 39/100 (39%)
Frame = +1
Query: 175 DLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNK 234
++P CP+ +MKD V S ++ + I ++ SK C T L D
Sbjct: 127 EVPSFFVCPISLEIMKDPVTVSTGI--TYDRESIEKWLFSKKNMTCPVTKQLLSDYTDLT 300
Query: 235 TLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIP 274
T + R++++ + + G R P PK P
Sbjct: 301 TPNHTLRRLIQAWCTLNASQG---------IERIPTPKPP 393
>BQ137239 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (13%)
Length = 1097
Score = 29.6 bits (65), Expect = 2.8
Identities = 15/26 (57%), Positives = 16/26 (60%)
Frame = +1
Query: 65 GAGRGFRRGAVGGRIGGGRGFGLERK 90
G G G R G GGR GGGR G ER+
Sbjct: 325 GGGWGGRGGGGGGRGGGGRRDGSERR 402
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 29.3 bits (64), Expect = 3.7
Identities = 16/41 (39%), Positives = 20/41 (48%), Gaps = 2/41 (4%)
Frame = +2
Query: 73 GAVGGRIGGGRGFGLERKTPPEG--YVCHRCKVSGHFIQHC 111
G GGR GGG G G ++ G C+ C GHF + C
Sbjct: 263 GGGGGRGGGGGGGGGRGRSGGGGSDLKCYXCGEPGHFARXC 385
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,795,379
Number of Sequences: 36976
Number of extensions: 250531
Number of successful extensions: 1845
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1728
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1815
length of query: 498
length of database: 9,014,727
effective HSP length: 100
effective length of query: 398
effective length of database: 5,317,127
effective search space: 2116216546
effective search space used: 2116216546
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126012.10


