

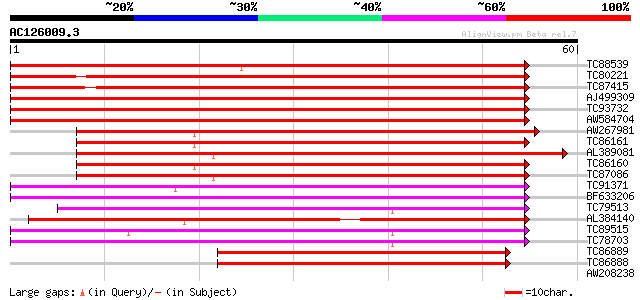
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.3 - phase: 0 /pseudo
(60 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88539 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 107 1e-24
TC80221 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 93 2e-20
TC87415 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 90 2e-19
AJ499309 weakly similar to GP|7670832|gb|A dicyanin {Lycopersico... 62 3e-11
TC93732 homologue to OMNI|NT01MC2635 Integral membrane protein o... 62 4e-11
AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor... 60 1e-10
AW267981 similar to PIR|H96745|H967 probable blue copper protein... 54 9e-09
TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 53 3e-08
AL389081 similar to SP|Q41001|BCP_P Blue copper protein precurso... 52 4e-08
TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 52 5e-08
TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precurs... 50 2e-07
TC91371 weakly similar to GP|14140127|emb|CAC39044. uclacyanin 3... 45 7e-06
BF633206 similar to GP|21553614|gb| blue copper protein putativ... 44 1e-05
TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin... 44 1e-05
AL384140 weakly similar to GP|2094888|pdb|2 Cucumber Basic Prote... 43 3e-05
TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains sim... 41 1e-04
TC78703 homologue to GP|13129456|gb|AAK13114.1 Putative retroele... 41 1e-04
TC86889 similar to PIR|S71274|S71274 blue copper-binding protein... 40 2e-04
TC86888 similar to PIR|S71274|S71274 blue copper-binding protein... 40 2e-04
AW208238 SP|P93328|NO16 Early nodulin 16 precursor (N-16). [Barr... 36 0.003
>TC88539 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (37%)
Length = 835
Score = 107 bits (266), Expect = 1e-24
Identities = 55/56 (98%), Positives = 55/56 (98%), Gaps = 1/56 (1%)
Frame = +3
Query: 1 MASSRVVLILSISMVLLSSVAIA-TDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MASSRVVLILSISMVLLSSVAIA TDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL
Sbjct: 105 MASSRVVLILSISMVLLSSVAIAATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 272
>TC80221 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (40%)
Length = 813
Score = 92.8 bits (229), Expect = 2e-20
Identities = 46/55 (83%), Positives = 50/55 (90%)
Frame = +1
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MA +RV IL+ISMVLLSSVA+A DHIVGDDKGWTVDF+YTQW QDKVFRVGDNL
Sbjct: 67 MALNRVA-ILAISMVLLSSVAMAADHIVGDDKGWTVDFNYTQWTQDKVFRVGDNL 228
>TC87415 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (31%)
Length = 710
Score = 89.7 bits (221), Expect = 2e-19
Identities = 43/55 (78%), Positives = 49/55 (88%)
Frame = +1
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
M SR + L+ISMVLLSSVA+A DH+VGD+KGWTVDF+YTQWAQDKVFRVGDNL
Sbjct: 67 MTYSRAIY-LAISMVLLSSVAMAADHVVGDEKGWTVDFNYTQWAQDKVFRVGDNL 228
>AJ499309 weakly similar to GP|7670832|gb|A dicyanin {Lycopersicon
esculentum}, partial (12%)
Length = 342
Score = 62.4 bits (150), Expect = 3e-11
Identities = 26/55 (47%), Positives = 39/55 (70%)
Frame = -3
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MA SRV+ + ++ + S++A+A D +VGD++GW + DY WA +KVFRVGD L
Sbjct: 310 MALSRVLFLFALIATIFSTMAVAKDFVVGDERGWKLGVDYQYWAANKVFRVGDTL 146
>TC93732 homologue to OMNI|NT01MC2635 Integral membrane protein of unknown
function {Magnetococcus sp. MC-1}, partial (2%)
Length = 443
Score = 62.0 bits (149), Expect = 4e-11
Identities = 26/55 (47%), Positives = 38/55 (68%)
Frame = +3
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MA SR + + ++ + S++A+A D +VGD+KGWT FDY W +KVFR+GD L
Sbjct: 54 MALSRALFLFALIASIFSTMAVAKDFVVGDEKGWTTLFDYQTWTANKVFRLGDTL 218
>AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor (Fib-H)
(H-fibroin). [Silk moth] {Bombyx mori}, partial (0%)
Length = 679
Score = 60.5 bits (145), Expect = 1e-10
Identities = 25/55 (45%), Positives = 38/55 (68%)
Frame = +1
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MA SR + + ++ + S++A+A D +VGD+ GWT+ DY WA +KVFR+GD L
Sbjct: 52 MALSRSLFLFALIATIFSTMAVAKDFVVGDESGWTLGVDYQAWAANKVFRLGDTL 216
>AW267981 similar to PIR|H96745|H967 probable blue copper protein T9N14.17
[imported] - Arabidopsis thaliana, partial (14%)
Length = 584
Score = 54.3 bits (129), Expect = 9e-09
Identities = 26/50 (52%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Frame = +3
Query: 8 LILSISMVLLS-SVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNLG 56
+I S ++LL+ A ATD VGD GW + DYT+WA K F+VGDNLG
Sbjct: 90 MIASFFVLLLAFPYAFATDFTVGDANGWNLGVDYTKWASGKTFKVGDNLG 239
>TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (17%)
Length = 736
Score = 52.8 bits (125), Expect = 3e-08
Identities = 26/49 (53%), Positives = 32/49 (65%), Gaps = 1/49 (2%)
Frame = +1
Query: 8 LILSISMVLLS-SVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
+I S ++LL+ A ATD VGD GWT DYT+WA K F+VGDNL
Sbjct: 112 MIASFFVLLLAFPYAFATDFTVGDANGWTQGVDYTKWASGKTFKVGDNL 258
>AL389081 similar to SP|Q41001|BCP_P Blue copper protein precursor. [Garden
pea] {Pisum sativum}, partial (50%)
Length = 491
Score = 52.4 bits (124), Expect = 4e-08
Identities = 25/53 (47%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Frame = +1
Query: 8 LILSISMVLLSSV-AIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNLGTMF 59
LIL +V+ +V +AT H VGD GW + DY WA DK F VGD+LG +
Sbjct: 25 LILGFFLVINVAVPTLATVHTVGDKSGWAIGSDYNTWASDKTFAVGDSLGKYY 183
>TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (34%)
Length = 918
Score = 52.0 bits (123), Expect = 5e-08
Identities = 25/49 (51%), Positives = 32/49 (65%), Gaps = 1/49 (2%)
Frame = +3
Query: 8 LILSISMVLLS-SVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
+I S ++LL+ A ATD VGD GW + DYT+WA K F+VGDNL
Sbjct: 132 MIASFFVLLLAFPYAFATDFTVGDANGWNLGVDYTKWASGKTFKVGDNL 278
>TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precursor. [Garden
pea] {Pisum sativum}, partial (87%)
Length = 1219
Score = 49.7 bits (117), Expect = 2e-07
Identities = 24/49 (48%), Positives = 30/49 (60%), Gaps = 1/49 (2%)
Frame = +1
Query: 8 LILSISMVLLSSV-AIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
LIL +V+ +V +AT H VGD GW + DY WA DK F VGD+L
Sbjct: 451 LILGFFLVINVAVPTLATVHTVGDKSGWAIGSDYNTWASDKTFAVGDSL 597
>TC91371 weakly similar to GP|14140127|emb|CAC39044. uclacyanin 3-like
protein {Oryza sativa}, partial (48%)
Length = 686
Score = 44.7 bits (104), Expect = 7e-06
Identities = 19/56 (33%), Positives = 34/56 (59%), Gaps = 1/56 (1%)
Frame = +1
Query: 1 MASSRVVLILSISMVL-LSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
MA S + ++L + + ++ +AT + VGD GW + +Y++W +K F VGD+L
Sbjct: 40 MAFSNIAMVLCFFLAINMALPTLATFYTVGDSLGWQIGVEYSKWTSEKTFVVGDSL 207
>BF633206 similar to GP|21553614|gb| blue copper protein putative
{Arabidopsis thaliana}, partial (56%)
Length = 478
Score = 44.3 bits (103), Expect = 1e-05
Identities = 17/55 (30%), Positives = 30/55 (53%)
Frame = +2
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
M +++L+ + L++ +AT H VG +GW D+ W+ + F+VGD L
Sbjct: 38 MGCKKMILLALLLATLITKEVLATQHNVGGSQGWDPSSDFDSWSSGQTFKVGDQL 202
>TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin-like
protein CASLP1 precursor {Capsicum annuum}, partial
(33%)
Length = 1093
Score = 43.9 bits (102), Expect = 1e-05
Identities = 21/54 (38%), Positives = 30/54 (54%), Gaps = 4/54 (7%)
Frame = +3
Query: 6 VVLILSISMVLLSSVAIATDHIVGDDKGWTVDFD----YTQWAQDKVFRVGDNL 55
V ++L+++ L T H+VGD GWT+ + YT WA +K F VGD L
Sbjct: 417 VFVLLAVAANLFHGSFAQTRHVVGDTTGWTIPTNGASFYTNWASNKTFTVGDTL 578
>AL384140 weakly similar to GP|2094888|pdb|2 Cucumber Basic Protein A Blue
Copper Protein, partial (92%)
Length = 441
Score = 42.7 bits (99), Expect = 3e-05
Identities = 21/55 (38%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Frame = +2
Query: 3 SSRVVLILSISMVLL--SSVAIATDHIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 55
SS ++++S+ +L+ S+++ A +HIVGD KGW+ F W K F+ GD L
Sbjct: 56 SSTSIILMSLLCMLVFHSNMSFAEEHIVGDGKGWS--FGVQNWPAGKTFKAGDTL 214
>TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains similarity to
nodulin~gene_id:K10D20.11 {Arabidopsis thaliana},
partial (43%)
Length = 854
Score = 40.8 bits (94), Expect = 1e-04
Identities = 23/60 (38%), Positives = 36/60 (59%), Gaps = 5/60 (8%)
Frame = +2
Query: 1 MASSRVVLILS-ISMVLLSSVAIATDHIVGDDKGWTVDFD----YTQWAQDKVFRVGDNL 55
+ S+++V LS ++L+ + A D IVG KGW+V D + QWA+ F+VGD+L
Sbjct: 59 LRSNKIVHALSWFCLMLMIHKSAAYDFIVGGQKGWSVPSDSNNPFNQWAEKSRFQVGDSL 238
>TC78703 homologue to GP|13129456|gb|AAK13114.1 Putative retroelement pol
polyprotein {Oryza sativa} [Oryza sativa (japonica
cultivar-group)], partial (2%)
Length = 1235
Score = 40.8 bits (94), Expect = 1e-04
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 5/60 (8%)
Frame = +3
Query: 1 MASSRVVLILSISMVLLSSVAIATDHIVGDDKGWTVDFD-----YTQWAQDKVFRVGDNL 55
MA+ L+L +VLL A + +VG KGW+ D Y QWA+ F+VGD+L
Sbjct: 75 MATIVQALVLFCLLVLLMHKGDAYEFVVGGQKGWSAPSDPNANPYNQWAEKSRFQVGDSL 254
>TC86889 similar to PIR|S71274|S71274 blue copper-binding protein 15K -
Arabidopsis thaliana, partial (14%)
Length = 688
Score = 40.0 bits (92), Expect = 2e-04
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +3
Query: 23 ATDHIVGDDKGWTVDFDYTQWAQDKVFRVGD 53
ATDHIVG ++GW +YT WA + VGD
Sbjct: 60 ATDHIVGANRGWNPGINYTLWANNHTIYVGD 152
>TC86888 similar to PIR|S71274|S71274 blue copper-binding protein 15K -
Arabidopsis thaliana, partial (14%)
Length = 697
Score = 40.0 bits (92), Expect = 2e-04
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +1
Query: 23 ATDHIVGDDKGWTVDFDYTQWAQDKVFRVGD 53
ATDHIVG ++GW +YT WA + VGD
Sbjct: 73 ATDHIVGANRGWNPGINYTLWANNHTIYVGD 165
>AW208238 SP|P93328|NO16 Early nodulin 16 precursor (N-16). [Barrel medic]
{Medicago truncatula}, partial (51%)
Length = 402
Score = 36.2 bits (82), Expect = 0.003
Identities = 21/64 (32%), Positives = 35/64 (53%), Gaps = 5/64 (7%)
Frame = +3
Query: 2 ASSRVVLILSISMVLLSSVAIATDHIVGDD-KGWTVDFD----YTQWAQDKVFRVGDNLG 56
+SS ++L++ SM LL S + +TD+++GD W V + +WA F VGD +
Sbjct: 33 SSSPILLMIIFSMWLLISHSESTDYLIGDSHNSWKVPLPSRRAFARWASAHEFTVGDTIR 212
Query: 57 TMFS 60
+S
Sbjct: 213KPYS 224
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,632,011
Number of Sequences: 36976
Number of extensions: 14549
Number of successful extensions: 118
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 113
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 113
length of query: 60
length of database: 9,014,727
effective HSP length: 36
effective length of query: 24
effective length of database: 7,683,591
effective search space: 184406184
effective search space used: 184406184
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 51 (24.3 bits)
Medicago: description of AC126009.3


