

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.5 + phase: 0
(652 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
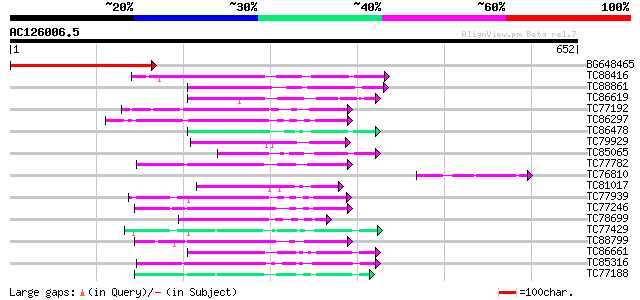
Score E
Sequences producing significant alignments: (bits) Value
BG648465 similar to GP|9955545|emb| ankyrin-repeat containing pr... 346 2e-95
TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110... 142 5e-34
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 80 3e-15
TC86619 similar to GP|16648824|gb|AAL25602.1 AT4g18950/F13C5_120... 74 2e-13
TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein... 72 5e-13
TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6 ... 71 1e-12
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 70 2e-12
TC79929 similar to GP|6729346|dbj|BAA89783.1 IRE {Arabidopsis th... 68 1e-11
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 67 2e-11
TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein k... 66 5e-11
TC76810 homologue to GP|17645764|emb|CAD19054. unnamed protein p... 65 8e-11
TC81017 weakly similar to GP|21632795|gb|AAL47185.1 phosphoinosi... 64 2e-10
TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {... 63 4e-10
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 61 1e-09
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 61 1e-09
TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120... 60 3e-09
TC88799 similar to PIR|C71408|C71408 probable protein kinase - A... 60 3e-09
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 60 3e-09
TC85316 weakly similar to PIR|E84707|E84707 probable protein kin... 59 4e-09
TC77188 similar to PIR|T04862|T04862 probable serine/threonine-s... 57 2e-08
>BG648465 similar to GP|9955545|emb| ankyrin-repeat containing protein
{Arabidopsis thaliana}, partial (9%)
Length = 787
Score = 346 bits (887), Expect = 2e-95
Identities = 166/169 (98%), Positives = 168/169 (99%)
Frame = +2
Query: 1 MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS 60
MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS
Sbjct: 260 MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS 439
Query: 61 VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS 120
VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS
Sbjct: 440 VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS 619
Query: 121 RGSQASSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRC 169
RGSQASSSGGCAPVIEVGVHQDLKLV+RIGEGRR+ VEMWSAVIGGGRC
Sbjct: 620 RGSQASSSGGCAPVIEVGVHQDLKLVKRIGEGRRS*VEMWSAVIGGGRC 766
>TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110
{Arabidopsis thaliana}, partial (52%)
Length = 2146
Score = 142 bits (357), Expect = 5e-34
Identities = 92/301 (30%), Positives = 150/301 (49%), Gaps = 5/301 (1%)
Frame = +3
Query: 141 QDLKLVRRIGEGRRAGVEMWSAVIGGGRCK----HQVAVKKVVLNEGMDLDWMLGKLEDL 196
+ LKL RIG G V W A + H+VA K + + + +L K +L
Sbjct: 135 ETLKLQHRIGRGPFGDV--WLATLHQSTEDYDEHHEVAAKMLHPIKEDHVKIVLKKFNEL 308
Query: 197 RRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLR-NEGRLTLEQVLRYGADIAR 255
+VC HG ++ +C++M GS+ ++ R G ++L VLRYG D+A+
Sbjct: 309 YLKCQGVSSVCWLHGISMLNGRICIIMKLYEGSIGDKLARLRNGWISLPDVLRYGIDLAQ 488
Query: 256 GVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCM 315
G++E HA G++ ++LKP N+L++ N A++ D G+ +L S+ + S +
Sbjct: 489 GILEHHAKGILVLNLKPCNVLINDNDQAILGDVGIPNLL-----------LGSSFVSSDI 635
Query: 316 ECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSA 375
+ SP+Y APE W+P + +S E+D+W FGCT+VEM TG+ PW G
Sbjct: 636 AQRLGSPNYMAPEQWKPEVRG---------PMSFETDSWGFGCTIVEMLTGSQPWYGCPV 788
Query: 376 EEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQE 435
IYR VV+ ++P + G+P + ++ C ++ RP +L F R L E
Sbjct: 789 GGIYRSVVEKHEKPH-----IPSGLPSPIENILSACFEYDMRNRPLMVDVLRAFKRSLNE 953
Query: 436 I 436
+
Sbjct: 954 L 956
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 79.7 bits (195), Expect = 3e-15
Identities = 60/233 (25%), Positives = 107/233 (45%), Gaps = 2/233 (0%)
Frame = +1
Query: 205 NVCTFHGAMKVDEGLCLVMDKCF-GSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
N+ ++G+ V + LC+ M+ GS+Q M + G +T V + I G+ LH+
Sbjct: 1207 NIVEYYGSEVVGDRLCIYMEYVHPGSLQKFMQDHCGVMTESVVRNFTRHILSGLAYLHST 1386
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
+ +K +NLL+DA+G ++D+G++ IL + S + + SP+
Sbjct: 1387 KTIHRDIKGANLLVDASGIVKLADFGVSKILTEKSYELS---------------LKGSPY 1521
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
+ APE K+ + ++ D WS GCT++EM TG PW+ + +V+
Sbjct: 1522 WMAPELMMAAMKN-----ETNPTVAMAVDIWSLGCTIIEMLTGKPPWSEFPGHQAMFKVL 1686
Query: 384 KAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLA-IFLRHLQE 435
P+ S G + +C Q P+ RP+ +L F+++L E
Sbjct: 1687 HRSPDIPKTLSPEG-------QDFLEQCFQRNPADRPSAAVLLTHPFVQNLHE 1824
>TC86619 similar to GP|16648824|gb|AAL25602.1 AT4g18950/F13C5_120
{Arabidopsis thaliana}, partial (77%)
Length = 1505
Score = 73.9 bits (180), Expect = 2e-13
Identities = 64/227 (28%), Positives = 104/227 (45%), Gaps = 5/227 (2%)
Frame = +3
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHA-- 262
NV F GA+ + +V + + ++ +G L +R+ DIARGV LH
Sbjct: 390 NVVQFLGAVTQSTPMMIVTEYLPKGDLRDFMKRKGALKPSTAVRFALDIARGVGYLHENK 569
Query: 263 -AGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLS 321
+ ++ L+PSN+L D +GH V+D+G++ +L + K + C S
Sbjct: 570 PSPIIHRDLEPSNILRDDSGHLKVADFGVSKLL-------------ANKEDKPLTCQETS 710
Query: 322 PHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQ 381
Y APE ++ + + D +SF L EM G P++ +E+ +
Sbjct: 711 CRYVAPEVFKQEEYDTKV------------DVFSFALILQEMIEGCPPFSAKRDDEV-PK 851
Query: 382 VVKAKKQPPQYASV--VGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
V +K++PP A + GI REL I EC P+KRPTF ++
Sbjct: 852 VYASKERPPFRAPIKRYSHGI-REL---IEECWNENPAKRPTFRQII 980
>TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein kinase
AtPK1/AtPK6 (EC 2.7.1.-). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (76%)
Length = 1992
Score = 72.4 bits (176), Expect = 5e-13
Identities = 68/266 (25%), Positives = 118/266 (43%)
Frame = +3
Query: 129 GGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDW 188
G + VG+ +D ++++ +G+G A V G ++K + E ++
Sbjct: 729 GNLMEIRRVGI-EDFEVLKVVGQGAFAKVYQVRKK-GTSEIYAMKVMRKDKIMEKNHAEY 902
Query: 189 MLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLR 248
M + E L T + V + + L LV+D G L ++G +
Sbjct: 903 MKAEREIL--TKIEHPFVVQLRYSFQTKYRLYLVLDFVNGGHLFFQLYHQGLFREDLARI 1076
Query: 249 YGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDS 308
Y A+I V LH+ G++ LKP N+L+DA+GH +++D+GLA ++ + ++ C +
Sbjct: 1077YAAEIVSAVSHLHSKGIMHRDLKPENILMDADGHVMLTDFGLAKKFEEST--RSNSLCGT 1250
Query: 309 AKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAI 368
A+ Y APE G G +D WS G L EM TG
Sbjct: 1251AE-------------YMAPEIIL------------GKGHDKAADWWSVGILLFEMLTGKP 1355
Query: 369 PWAGLSAEEIYRQVVKAKKQPPQYAS 394
P+ G + ++I +++VK K + P + S
Sbjct: 1356PFCGGNCQKIQQKIVKDKIKLPGFLS 1433
>TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6
{Arabidopsis thaliana}, partial (90%)
Length = 2043
Score = 70.9 bits (172), Expect = 1e-12
Identities = 72/287 (25%), Positives = 126/287 (43%), Gaps = 3/287 (1%)
Frame = +2
Query: 111 DQKLDCRKNSRGSQASSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCK 170
++K ++S S SS+GG A + G ++ L R +GEG A V+ + G
Sbjct: 371 EKKKMTSRSSHISGGSSNGGRA---KAGKYE---LGRTLGEGNFAKVKFARHIETGDHVA 532
Query: 171 HQVAVKKVVLNEGM--DLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFG 228
++ K+ +L M + + ++ +R NV H + + +VM+ G
Sbjct: 533 IKILDKEKILKHKMIRQIKQEISTMKLIRHP-----NVIRMHEVIANRSKIFIVMELVTG 697
Query: 229 SVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDY 288
+ + GRL ++ +Y + V H+ GV LKP NLLLD NG VSD+
Sbjct: 698 GELFDKIARSGRLKEDEARKYFQQLICAVDYCHSRGVCHRDLKPENLLLDTNGTLKVSDF 877
Query: 289 GLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGIS 348
GL+ + ++ + +H+ +P+Y APE + N +D I
Sbjct: 878 GLSALPQQVR--------EDGLLHTTCG----TPNYVAPEVIQ------NKGYDGAI--- 994
Query: 349 PESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAK-KQPPQYAS 394
+D WS G L + G +P+ + +Y+++ KA PP ++S
Sbjct: 995 --ADLWSCGVILFVLMAGYLPFEEDNLVALYKKIFKADFTCPPWFSS 1129
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 70.5 bits (171), Expect = 2e-12
Identities = 55/222 (24%), Positives = 90/222 (39%)
Frame = +2
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ + G+ +E L + ++ G ++L+ G + Y I G+ LH
Sbjct: 1154 NIVQYLGSELGEESLSVYLEYVSGGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHGRN 1333
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHY 324
V +K +N+L+D NG ++D+G+A + + M SP++
Sbjct: 1334 TVHRDIKGANILVDPNGEIKLADFGMAKHITSAAS---------------MLSFKGSPYW 1468
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVK 384
APE V + N G S D WS GCTL+EM PW+ ++
Sbjct: 1469 MAPE----VVMNTN-------GYSLPVDIWSLGCTLIEMAASKPPWSQYEGVAAIFKIGN 1615
Query: 385 AKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+K P ++ + + I CLQ PS RPT +L
Sbjct: 1616 SKDMP-----IIPEHLSNDAKNFIMLCLQRDPSARPTAQKLL 1726
>TC79929 similar to GP|6729346|dbj|BAA89783.1 IRE {Arabidopsis thaliana},
partial (23%)
Length = 1383
Score = 67.8 bits (164), Expect = 1e-11
Identities = 53/200 (26%), Positives = 88/200 (43%), Gaps = 16/200 (8%)
Frame = +1
Query: 209 FHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCM 268
F+ + E L LVM+ G MLRN G L + Y A++ + LH+ +V
Sbjct: 7 FYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHR 186
Query: 269 SLKPSNLLLDANGHAVVSDYGLATI--------LKKPSCW--------KARPECDSAKIH 312
LKP NLL+ +GH ++D+GL+ + L P+ + + +P S +
Sbjct: 187 DLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREA 366
Query: 313 SCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAG 372
+ I+ +P Y APE G+G +D WS G L E+ G P+
Sbjct: 367 RQQQSIVGTPDYLAPEILL------------GMGHGTTADWWSVGVILYELLVGIPPFNA 510
Query: 373 LSAEEIYRQVVKAKKQPPQY 392
A++I+ ++ Q P++
Sbjct: 511 DHAQQIFDNIINRDIQWPKH 570
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 67.0 bits (162), Expect = 2e-11
Identities = 52/187 (27%), Positives = 80/187 (41%)
Frame = +2
Query: 240 RLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSC 299
+L QV Y I G+ LH +V +K +N+L+DANG V+D+GL+ +K
Sbjct: 8 KLRDSQVSAYTRQILHGLKYLHDRNIVHRDIKCANILVDANGSVKVADFGLS*AIKLND- 184
Query: 300 WKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCT 359
+ SC + + APE K L +D WS GCT
Sbjct: 185 -----------VKSCQG----TAFWMAPEVVRGKVKGYGL----------PADIWSLGCT 289
Query: 360 LVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKR 419
++EM TG +P+A + ++ K + P V + R+ I +CL+ P R
Sbjct: 290 VLEMLTGQVPYAPMECISAMFRIGKGELPP------VPDTLSRDARDFILQCLKVNPDDR 451
Query: 420 PTFNAML 426
PT +L
Sbjct: 452 PTAAQLL 472
>TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein kinase PKS6
{Arabidopsis thaliana}, partial (77%)
Length = 1236
Score = 65.9 bits (159), Expect = 5e-11
Identities = 64/248 (25%), Positives = 104/248 (41%)
Frame = +2
Query: 147 RRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNV 206
+ IGEG A V++ V G ++ + VL M +D + ++ ++ + NV
Sbjct: 203 KTIGEGSFAKVKLAKNVENGDYVAIKILDRNHVLKHNM-MDQLKREISAMKIINH--PNV 373
Query: 207 CTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVV 266
M + +V++ G + + GRL ++ Y + V H+ GV
Sbjct: 374 IKIFEVMASKTKIYIVLELVNGGELFDKIATHGRLKEDEARSYFQQLINAVDYCHSRGVY 553
Query: 267 CMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTA 326
LKP NLLLD NG VSD+GL+T S + + +P+Y A
Sbjct: 554 HRDLKPENLLLDTNGVLKVSDFGLSTY--------------SQQEDELLRTACGTPNYVA 691
Query: 327 PEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAK 386
PE LN D G + SD WS G L + G +P+ + +YR++ +A
Sbjct: 692 PEV-------LN---DRGY-VGSSSDIWSCGVILFVLMAGYLPFDEPNLIALYRKIGRAD 838
Query: 387 KQPPQYAS 394
+ P + S
Sbjct: 839 FKCPSWFS 862
>TC76810 homologue to GP|17645764|emb|CAD19054. unnamed protein product
{Glycine max}, partial (98%)
Length = 1439
Score = 65.1 bits (157), Expect = 8e-11
Identities = 42/133 (31%), Positives = 68/133 (50%)
Frame = +2
Query: 469 QDPNRLHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAEL 528
+D + +H S GD+ G++ LA A ++E ++++G+TALH AC G +
Sbjct: 749 EDESIVHHTASVGDIEGLKAALASGADKDE----------EDSEGRTALHFACGYGEVKC 898
Query: 529 VETILDYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGPSVAHV 588
+ +L+ A VD LDK+ + L +A G ECV L++ A VT + DG P V
Sbjct: 899 AQALLE-AGAKVDALDKNKNTALHYAAGYGRKECVALLLENGAAVTLQNMDGKTP--IDV 1069
Query: 589 CAYHGQPDCMREL 601
+ Q D ++ L
Sbjct: 1070AKLNNQDDVLQLL 1108
Score = 38.9 bits (89), Expect = 0.006
Identities = 20/71 (28%), Positives = 34/71 (47%)
Frame = +2
Query: 579 DGLGPSVAHVCAYHGQPDCMRELLLAGADPNAVDDEGESVLHRAIAKKFTDCALVIVENG 638
D G + H +G+ C + LL AGA +A+D + LH A +C +++ENG
Sbjct: 842 DSEGRTALHFACGYGEVKCAQALLEAGAKVDALDKNKNTALHYAAGYGRKECVALLLENG 1021
Query: 639 GCRSMAISNSK 649
++ + K
Sbjct: 1022AAVTLQNMDGK 1054
>TC81017 weakly similar to GP|21632795|gb|AAL47185.1
phosphoinositide-dependent protein kinase-1 beta {Mus
musculus}, partial (32%)
Length = 721
Score = 63.5 bits (153), Expect = 2e-10
Identities = 50/179 (27%), Positives = 80/179 (43%), Gaps = 11/179 (6%)
Frame = +2
Query: 216 DEG-LCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSN 274
DEG L V+D C G +L+ G ++ YGA I + +H+ GV+ LKP N
Sbjct: 209 DEGSLYYVLDLCNGGELLGVLKKTGTFDVDCTRFYGAQILDAIDYMHSRGVIHRDLKPEN 388
Query: 275 LLLDANGHAVVSDYGLATILKKP-------SCWKARPECDS---AKIHSCMECIMLSPHY 324
+LLD + H ++D+G A +LK P + PE S A S + + Y
Sbjct: 389 VLLDDHMHVKITDFGTAKLLKDPRDSQTPGDGGRGLPEGSSRSEAGDDSRAASFVGTAEY 568
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
+PE + S N SD W+FGC + ++ G P+ + ++++V
Sbjct: 569 VSPE----LLTSKN--------ACKASDLWAFGCIIYQLLAGRPPFKAATEYLTFQKIV 709
>TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {Arabidopsis
thaliana}, partial (70%)
Length = 1353
Score = 62.8 bits (151), Expect = 4e-10
Identities = 68/262 (25%), Positives = 109/262 (40%), Gaps = 5/262 (1%)
Frame = +1
Query: 137 VGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDL 196
VG+++ + R IGEG A V+ G ++ K+ VL M E +
Sbjct: 544 VGIYE---VGRTIGEGTFAKVKFARNSETGEAVALKILDKEKVLKHKM--------AEQI 690
Query: 197 RRTSMWCR-----NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGA 251
+R + NV + M + +V++ G + + N GR+ + RY
Sbjct: 691 KREIATMKLIKHPNVVRLYEVMGSRTKIYIVLEFVTGGELFDKIVNHGRMGEPEARRYFQ 870
Query: 252 DIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKI 311
+ V H+ GV LKP NLLLDA G+ VSD+GL+ + ++ D +
Sbjct: 871 QLINVVDYCHSRGVYHRDLKPENLLLDAYGNLKVSDFGLSALSQQVR--------DDGLL 1026
Query: 312 HSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWA 371
H+ +P+Y APE LN DG +D WS G L + G +P+
Sbjct: 1027HTTCG----TPNYVAPEV-------LNDRGYDG----ATADLWSCGVILFVLVAGYLPFD 1161
Query: 372 GLSAEEIYRQVVKAKKQPPQYA 393
+ E+Y+++ A P A
Sbjct: 1162DPNLMELYKKISSADFTCPPVA 1227
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 61.2 bits (147), Expect = 1e-09
Identities = 58/251 (23%), Positives = 102/251 (40%)
Frame = +3
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWC 203
+L + +G G A V ++I G +V K ++ M+ ++ +++ +RR
Sbjct: 90 QLTKFLGRGNFAKVYQARSLIDGSTVAVKVIDKSKTVDASME-PRIVREIDAMRRLQNH- 263
Query: 204 RNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
N+ H + + L++D G L + RY + + H
Sbjct: 264 PNILKIHEVLATKTKIYLIVDFAGGGELFLKLARRRKFPEALARRYFQQLVSALCFCHRN 443
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
G+ LKP NLLLDA G+ VSD+GL+ + ++ C +P
Sbjct: 444 GIAHRDLKPQNLLLDAEGNLKVSDFGLSALPEQLKNGLLHTACG-------------TPA 584
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
YTAPE L DG ++DAWS G L + G +P+ + + +++
Sbjct: 585 YTAPEI-------LGRIGYDG----SKADAWSCGVILYVLLAGHLPFDDSNIPAMCKRIT 731
Query: 384 KAKKQPPQYAS 394
+ Q P++ S
Sbjct: 732 RRDYQFPEWIS 764
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 61.2 bits (147), Expect = 1e-09
Identities = 41/177 (23%), Positives = 81/177 (45%), Gaps = 1/177 (0%)
Frame = +3
Query: 195 DLRRTSMWCRNVCTFHGAMKVDEG-LCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADI 253
++ R + C NV +HG+ + G +C++M+ L+ G + ++ DI
Sbjct: 345 NILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDI 524
Query: 254 ARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHS 313
G+ LHA + +KPSN+L++ ++D+G++ + + E ++ + +
Sbjct: 525 LNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTL------EACNSYVGT 686
Query: 314 CMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPW 370
C Y +PE ++P ++ + G S +D WS G TL E+ G P+
Sbjct: 687 CA--------YMSPERFDP-----EVYGGNYNGFS--ADIWSLGLTLFELYVGYFPF 812
>TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120
{Arabidopsis thaliana}, partial (79%)
Length = 1869
Score = 60.1 bits (144), Expect = 3e-09
Identities = 75/304 (24%), Positives = 121/304 (39%), Gaps = 8/304 (2%)
Frame = +3
Query: 133 PVIEVGVHQ---DLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWM 189
P+ EV H ++ R +G+G A V + G +V K V EGM
Sbjct: 231 PMEEVKAHMLFGKYEMGRVLGKGTFAKVYYAKEISTGEGVAIKVVDKDKVKKEGM----- 395
Query: 190 LGKLEDLRRTSMWCR-----NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLE 244
+E ++R R N+ M + VM+ G E + +G+ E
Sbjct: 396 ---MEQIKREISVMRLVKHPNIVNLKEVMATKTKILFVMEYARGGELFEKVA-KGKFKEE 563
Query: 245 QVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARP 304
+Y + V H+ GV LKP NLLLD N + VSD+GL+ A P
Sbjct: 564 LARKYFQQLISAVDYCHSRGVSHRDLKPENLLLDENENLKVSDFGLS----------ALP 713
Query: 305 ECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMC 364
E + + +P Y APE K+ + F ++D WS G L +
Sbjct: 714 E--HLRQDGLLHTQCGTPAYVAPEVVR--KRGYSGF---------KADTWSCGVILYALL 854
Query: 365 TGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNA 424
G +P+ + +Y +V K + Q P + S E ++I + L P +R T ++
Sbjct: 855 AGFLPFQHENLISMYNKVFKEEYQFPPWFS-------PESKRLISKILVADPERRITISS 1013
Query: 425 MLAI 428
++ +
Sbjct: 1014IMNV 1025
>TC88799 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (57%)
Length = 1648
Score = 60.1 bits (144), Expect = 3e-09
Identities = 59/254 (23%), Positives = 104/254 (40%), Gaps = 3/254 (1%)
Frame = +1
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDW---MLGKLEDLRRTS 200
+L R +G G A V +++ + VAVK + ++ +D ++ +++ +RR
Sbjct: 130 QLTRLLGRGSFAKVYKAHSLLDNA---NTVAVKIIDKSKNVDAAMEPRIIREIDAMRRLQ 300
Query: 201 MWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVEL 260
N+ H M + L+++ G L GR + Y + +
Sbjct: 301 HH-PNILKIHEVMATKTKIHLIVEFAAGGELFSALSRRGRFSESTARFYFQQLVSALRFC 477
Query: 261 HAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIML 320
H GV LKP NLLLDA+G+ VSD+GL+ + + C
Sbjct: 478 HRNGVAHRDLKPQNLLLDADGNLKVSDFGLSALPEHLKDGLLHTACG------------- 618
Query: 321 SPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYR 380
+P YTAPE + G ++DAWS G L + G +P+ L+ + +
Sbjct: 619 TPAYTAPE----------ILRRSGGYDGSKADAWSCGLILYVLLAGYLPFDDLNIPAMCK 768
Query: 381 QVVKAKKQPPQYAS 394
++ + + P + S
Sbjct: 769 KISRRDYRFPDWVS 810
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 59.7 bits (143), Expect = 3e-09
Identities = 52/222 (23%), Positives = 90/222 (40%)
Frame = +2
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ H + + VM+ G + N+GR + + R + V H+ G
Sbjct: 482 NIVRLHEVLATKTKIYFVMEFAKGGELFAKIANKGRFSEDLSRRLFQQLISAVGYCHSHG 661
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHY 324
V LKP NLLLD G+ VSD+GL+ + + +I + + +P Y
Sbjct: 662 VFHRDLKPENLLLDDKGNLKVSDFGLSAVK------------EQVRIDGMLHTLCGTPAY 805
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVK 384
APE K+ DG + D WS G L + G +P+ + +YR++
Sbjct: 806 VAPEIL--AKRGY-----DGAKV----DIWSCGIILFVLVAGYLPFNDPNLMVMYRKIYS 952
Query: 385 AKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+ + P++ S +L + + + P R T + +L
Sbjct: 953 GEFKCPRWFS-------PDLKRFLSRLMDTNPETRITVDEIL 1057
>TC85316 weakly similar to PIR|E84707|E84707 probable protein kinase
[imported] - Arabidopsis thaliana, partial (66%)
Length = 1528
Score = 59.3 bits (142), Expect = 4e-09
Identities = 62/280 (22%), Positives = 113/280 (40%)
Frame = +1
Query: 147 RRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNV 206
R +G G A V + + G +V KK ++N G + ++ LRR + N+
Sbjct: 274 RLLGVGASAKVYHATNIETGKSVAVKVISKKKLINNGEFAANIEREISILRR--LHHPNI 447
Query: 207 CTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVV 266
+ + V++ E + + +LT + RY + V H+ GV
Sbjct: 448 IDLFEVLASKSKIYFVVEFAEAGELFEEVAKKEKLTEDHARRYFRQLISAVKHCHSRGVF 627
Query: 267 CMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTA 326
LK NLLLD N + V+D+GL+ + + RP+ + + +P Y A
Sbjct: 628 HRDLKLDNLLLDENDNLKVTDFGLSAVKN-----QIRPD-------GLLHTVCGTPSYVA 771
Query: 327 PEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAK 386
PE KK ++D WS G L + G +P+ + +YR++ + +
Sbjct: 772 PEIL--AKKGYE---------GAKADVWSCGVVLFTVTAGYLPFNDYNVTVLYRKIYRGQ 918
Query: 387 KQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+ P++ S +L ++ L P R + + +L
Sbjct: 919 FRFPKWMSC-------DLKNLLSRMLDTNPKTRISVDEIL 1017
>TC77188 similar to PIR|T04862|T04862 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F28A21.110 - Arabidopsis
thaliana, partial (79%)
Length = 1881
Score = 57.0 bits (136), Expect = 2e-08
Identities = 68/277 (24%), Positives = 109/277 (38%), Gaps = 1/277 (0%)
Frame = +1
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWC 203
+L + +G G A V + + G ++ K +L G+ + + ++ LRR
Sbjct: 193 ELGKLLGHGTFAKVHLAKNLKTGESVAIKIISKDKILKSGL-VSHIKREISILRRVRH-- 363
Query: 204 RNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
N+ M + VM+ G + +GRL E +Y + V HA
Sbjct: 364 PNIVQLFEVMATKTKIYFVMEYVRGGELFNKVA-KGRLKEEVARKYFQQLICAVGFCHAR 540
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
GV LKP NLLLD G+ VSD+GL+ + D K +P
Sbjct: 541 GVFHRDLKPENLLLDEKGNLKVSDFGLSAV------------SDEIKQDGLFHTFCGTPA 684
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAG-LSAEEIYRQV 382
Y APE L+ DG + D WS G L + G +P+ + +Y+++
Sbjct: 685 YVAPEV-------LSRKGYDGAKV----DIWSCGVVLFVLMAGYLPFHDPNNVMAMYKKI 831
Query: 383 VKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKR 419
K + + P++ S EL ++ L KP R
Sbjct: 832 YKGEFRCPRWFS-------PELVSLLTRLLDIKPQTR 921
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,299,628
Number of Sequences: 36976
Number of extensions: 349371
Number of successful extensions: 2828
Number of sequences better than 10.0: 480
Number of HSP's better than 10.0 without gapping: 2533
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2702
length of query: 652
length of database: 9,014,727
effective HSP length: 102
effective length of query: 550
effective length of database: 5,243,175
effective search space: 2883746250
effective search space used: 2883746250
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126006.5


