

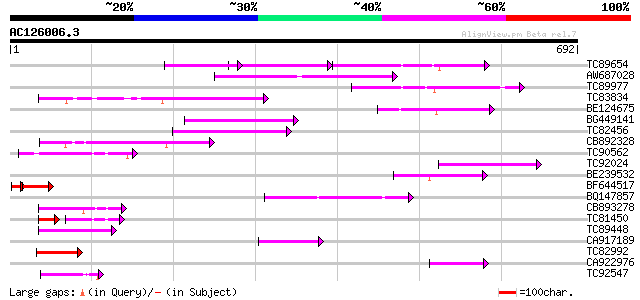
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.3 + phase: 0
(692 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 97 2e-37
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 146 3e-35
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 122 6e-28
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 115 6e-26
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 115 6e-26
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 114 1e-25
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 114 2e-25
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 92 5e-19
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 84 1e-16
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 80 3e-15
BE239532 79 8e-15
BF644517 66 1e-12
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 71 2e-12
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 68 1e-11
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 55 6e-11
TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43... 60 3e-09
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 60 4e-09
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 58 1e-08
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 54 2e-07
TC92547 similar to PIR|A56235|A56235 transcription activator Maf... 52 1e-06
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 97.1 bits (240), Expect(2) = 2e-37
Identities = 55/198 (27%), Positives = 93/198 (46%), Gaps = 6/198 (3%)
Frame = +1
Query: 394 DSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGRI 453
+ W + ++ ++ W VY+R F + +E N ++ + L + +
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 454 VHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNL 513
V E +AD+E+ + P L+ +PM QA ++YT K F +FQEE + +
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTP-SPMEKQAASLYTRKMFMKFQEELVGTMANPATKI 1053
Query: 514 KEGLYVVTNY------DNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIK 567
++ +T Y +N V N + K +C C+ FE GI+C H L V N+
Sbjct: 1054EDS-GTITTYRVAKFGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFRAKNVL 1230
Query: 568 LIPQHYILKRWTRDARLG 585
+P HY+LKRWT +A+ G
Sbjct: 1231TLPSHYVLKRWTMNAKTG 1284
Score = 77.8 bits (190), Expect(2) = 2e-37
Identities = 38/126 (30%), Positives = 68/126 (53%)
Frame = +3
Query: 268 LGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAI 327
LG+ + +N + L+ +E+ PS+ WLF+T+L+A+ G P ++ TD D + A+
Sbjct: 330 LGLIIMVN----LFYLAVLLILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAV 497
Query: 328 SVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLV 387
+ V+P T H C W I ++ + HLYQ + F ++F+ C+ EF + W LL
Sbjct: 498 AQVLPPTRHRFCKWSIFRENRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLE 677
Query: 388 EHNVSE 393
+ + +
Sbjct: 678 RYYIMD 695
Score = 70.1 bits (170), Expect = 3e-12
Identities = 36/95 (37%), Positives = 52/95 (53%)
Frame = +2
Query: 190 YLQTRRMRSLMYGEVGALLMHFKRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCS 249
++ + R R+L G L + ++EN +F+Y Q DV+ N+FWAD +Y
Sbjct: 83 HMSSTRQRTLGGGGHQVLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYF 262
Query: 250 GDVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFG 284
GD + FDTTY TN+ P F G N+H Q V+FG
Sbjct: 263 GDTVIFDTTYKTNQYRVPFASFTGFNHHGQPVLFG 367
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 146 bits (368), Expect = 3e-35
Identities = 78/224 (34%), Positives = 121/224 (53%), Gaps = 1/224 (0%)
Frame = +1
Query: 251 DVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGE 310
DV+ FDTTY NK PL +F G N+H Q ++FG LL DETI S++W+ FL+ M +
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 311 KPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACID 370
PK ++TD D +M +AI V P H LC W + +NAQ ++ K + F F +
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENI-----KKTPFLEGFRKAMY 345
Query: 371 LHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFN 430
+ +F + W+ L+ ++ + ++W+ + K WA Y+R F +R+T E+ N
Sbjct: 346 SNFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAIN 525
Query: 431 AELKNYLKSDLNLVQFFSHF-GRIVHGIRNNESEADYESRHKLP 473
A +K Y +S ++ F R G RNNE AD++S+ P
Sbjct: 526 AIVKTYSRSQRQNF*IYA*FLNRF*EGYRNNELVADFKSKFTEP 657
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 122 bits (305), Expect = 6e-28
Identities = 74/216 (34%), Positives = 110/216 (50%), Gaps = 5/216 (2%)
Frame = +2
Query: 418 AGMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKM 477
AG + Q SES N+ Y+ + L +F + I+ + E AD+++ HK P LK
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 478 KRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNLKEG-----LYVVTNYDNNKERMVI 532
+P Q IYT F++FQ E G C ++ G Y+V +Y+ ++E +V
Sbjct: 182 P-SPWEKQMSTIYTHAIFKKFQVEVLGVAG-CQSRIEVGDGTAVRYIVQDYEKDEEFLVT 355
Query: 533 GNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKG 592
+ +V+C CR FE G LC HAL VL +P HYI+KRWT+DA++ +
Sbjct: 356 WKELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAKIREPAADRT 535
Query: 593 QHIELDTKAHFMRRYNDLCPQMIKLINKASKSHETY 628
Q I+ +RYNDLC + I L + S S E+Y
Sbjct: 536 QRIQTRP-----QRYNDLCKRSIALSEEGSSSEESY 628
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 115 bits (288), Expect = 6e-26
Identities = 86/292 (29%), Positives = 137/292 (46%), Gaps = 12/292 (4%)
Frame = +3
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYG----NKSKKDGILTSRRFTCFKEGKRG 91
P L MEFES AY +Y Y++ +GF +R + N +K G + C +G +
Sbjct: 213 PALAMEFESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEKYGAV----LCCSSQGFKR 380
Query: 92 VDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHR 151
++L KE TRTGC A + + L ++++ + +HNH+L + H+
Sbjct: 381 TKDVNNLRKE----TRTGCPAMIRMKLVES-QRWRICEVTLEHNHVLGA-------KIHK 524
Query: 152 QISESRACQVVAADESRLKRKDFQEYVFKQDGGI-------DDVGYLQTRRMRSLMYGEV 204
I ++ + ++D K + V +G DD + + +L G+
Sbjct: 525 SIKKN---SLPSSDAEGKTIKVYHALVIDTEGNDNLNSNARDDRAFSKYSNKLNLRKGDT 695
Query: 205 GALLMHFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNK 263
A+ R Q NP+F+Y + E + N W DA+ G DVI FD TY+ NK
Sbjct: 696 QAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYLVNK 875
Query: 264 DYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTL 315
PL VG+N+H Q V+ G LL E I S++WLF T++K + P+T+
Sbjct: 876 YEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 115 bits (288), Expect = 6e-26
Identities = 67/146 (45%), Positives = 83/146 (55%), Gaps = 4/146 (2%)
Frame = +2
Query: 450 FGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTC 509
F RIV R E EA E + LPKL M +L A YTP+ FE FQ+ YE+ L
Sbjct: 2 FERIVEEQRYKEIEASDEMKGCLPKL-MGNVVVLKHASVAYTPRAFEVFQQRYEKSLNVI 178
Query: 510 VKNLKEGLYV----VTNYDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMN 565
V K Y+ V Y + ++ V + D V C C KFE G LCSHALKVLD N
Sbjct: 179 VNQHKRDGYLFEYKVNTYGHARQYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLDNRN 358
Query: 566 IKLIPQHYILKRWTRDARLGSNHDLK 591
IK++P YILKRWT+DARLG+ ++K
Sbjct: 359 IKVVPSRYILKRWTKDARLGNIREIK 436
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 114 bits (285), Expect = 1e-25
Identities = 52/139 (37%), Positives = 80/139 (57%)
Frame = +3
Query: 214 QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPLGVFVG 273
+ ++P+F +++ +D ++ N+ W+ A I Y GD + FDTT+ PLG++VG
Sbjct: 267 KDKDPNFKFEYTLDANNRLENIAWSYASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVG 446
Query: 274 LNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAISVVMPQ 333
+NN+ FG LL DET+ SF W + FL M G+ P+T+LTDQ+ + +A+S MP
Sbjct: 447 INNYGMPCFFGCVLLRDETVRSFSWAIKAFLGFMNGKAPQTILTDQNICLKEALSAEMPM 626
Query: 334 TFHGLCTWRIRENAQTHVN 352
T H C W I + VN
Sbjct: 627 TKHAFCIWMIVAKFPSWVN 683
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 114 bits (284), Expect = 2e-25
Identities = 59/146 (40%), Positives = 83/146 (56%), Gaps = 1/146 (0%)
Frame = +2
Query: 199 LMYGEVGALLMHFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDT 257
L G+V A+ F R Q +N FY MD + I N+FWADA+ Y G+VIT DT
Sbjct: 704 LWRGDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDT 883
Query: 258 TYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLT 317
TY+T+K PL FVG+N+H Q ++ G +L + + WLF +L+ M G P ++T
Sbjct: 884 TYLTSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIIT 1063
Query: 318 DQDEAMAKAISVVMPQTFHGLCTWRI 343
++D+AM AI V P+ H C W I
Sbjct: 1064EEDKAMKNAIEVAFPKARHRWCLWHI 1141
Score = 38.5 bits (88), Expect = 0.009
Identities = 30/108 (27%), Positives = 47/108 (42%), Gaps = 2/108 (1%)
Frame = +1
Query: 33 DYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGV 92
D P M F SE +Y Y+ R+GFG+ + +K+ DG + FT R
Sbjct: 199 DEEPRTGMTFRSEEEVIRYYMNYANRVGFGVTK-ISSKNADDG---KKYFTLACNCARRY 366
Query: 93 DKRDHLTKEGRVETRTGCDARM--VISLDRKIGKYKVVDFVAQHNHLL 138
+ + ++T C AR+ ++LD G + V +HNH L
Sbjct: 367 VSTSKNPSKQYLTSKTQCRARLNACVALD---GSSTISRIVLEHNHEL 501
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 92.4 bits (228), Expect = 5e-19
Identities = 60/227 (26%), Positives = 115/227 (50%), Gaps = 13/227 (5%)
Frame = +1
Query: 37 HLEMEFESEAAAYEFYNKYSRRIGFGIRRE----YGNKSKKDGILTSRRFTCFKEGKRGV 92
+L S+ AY+ Y +++ + GF +R+ Y N+ KK + + F C K+G +
Sbjct: 157 YLGQVVNSKDEAYDLYQEHAFKTGFSVRKGKELYYDNEKKKTRL---KDFYCSKQGFKN- 324
Query: 93 DKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQ 152
++ D R ++RT C A + ++ ++ G +KV + HNH P + RS R
Sbjct: 325 NEPDGEVAYKRADSRTNCLAMVRFNVTKE-GVWKVTKLILDHNHEFVPLQQRYLLRSMRN 501
Query: 153 ISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDVG--------YLQTRRMRSLMYGEV 204
+S + ++ + ++ + Y+ K+ GG+D G Y+ T +++ + G+
Sbjct: 502 MSNLKEDRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMHNYVSTGKLKFIEAGDA 681
Query: 205 GALLMHFK-RQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSG 250
+LL H + +Q+++ FYY Q+D E ++ NVFW D + + DY +G
Sbjct: 682 QSLLNHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKSVVDYIATG 822
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 84.3 bits (207), Expect = 1e-16
Identities = 52/153 (33%), Positives = 81/153 (51%), Gaps = 8/153 (5%)
Frame = +2
Query: 11 GVEKLGNNIDVNIVQNGEESVVDYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNK 70
GV+++ + + V V + P++ EF SEA A+ FYN Y+ R+GF IR ++
Sbjct: 263 GVDQIASAVPVTSVMRTIDE-----PYIGQEFVSEAEAHAFYNSYATRVGFVIRVSKLSR 427
Query: 71 SKKDGILTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKI--GKYKVV 128
S++DG + R C KEG R DKR+ + ++ R ETR GC A +++ RK+ G + +
Sbjct: 428 SRRDGSVIGRALVCNKEGFRMPDKREKIVRQ-RAETRVGCRAMIMV---RKLNSGLWSIT 595
Query: 129 DFVAQHNHLLEPPG------YFHTPRSHRQISE 155
FV +H H L P Y P H ++ E
Sbjct: 596 KFVKEHTHPLTPGKSRRDFVYEQYPSGHHRVRE 694
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 80.1 bits (196), Expect = 3e-15
Identities = 40/126 (31%), Positives = 64/126 (50%)
Frame = +3
Query: 524 DNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDAR 583
+++K V N+++ K C C+ FE G+LC H L V V N+ +P HYILKRWT++A+
Sbjct: 39 EDHKAYNVRFNVLEMKATCSCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWTKNAK 218
Query: 584 LGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASKSHETYTFLSKVFEESDKIID 643
+ H RYN L + K ++K + S ETY +E+ K +
Sbjct: 219 SNVSLQEHSSHAYTYYLESHTVRYNTLRHEAFKFVDKGASSPETYDVAKDALQEAAKRVA 398
Query: 644 DMLAKK 649
++ K+
Sbjct: 399 QVMRKE 416
>BE239532
Length = 645
Score = 78.6 bits (192), Expect = 8e-15
Identities = 44/121 (36%), Positives = 62/121 (50%), Gaps = 6/121 (4%)
Frame = +3
Query: 469 RHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVK------NLKEGLYVVTN 522
R P L + + L A +YT K F F EY E G N V+ N
Sbjct: 3 RQSAPGLVINISETLRHASTVYTHKGFRIFLNEYLEGTGGSTSIEISQSNNLSNHEVMLN 182
Query: 523 YDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDA 582
+ NK+ V + + K+ C CRKF++ GILCSHAL++ ++ I IP Y LKRW+++A
Sbjct: 183 HKPNKKYAVTFDSSNMKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLKRWSKNA 362
Query: 583 R 583
R
Sbjct: 363 R 365
>BF644517
Length = 227
Score = 65.9 bits (159), Expect(2) = 1e-12
Identities = 31/38 (81%), Positives = 32/38 (83%)
Frame = +3
Query: 16 GNNIDVNIVQNGEESVVDYIPHLEMEFESEAAAYEFYN 53
G VNIVQNGEE+VVDYIPHLEME ESEA AYEFYN
Sbjct: 114 GTIFSVNIVQNGEETVVDYIPHLEMESESEAVAYEFYN 227
Score = 25.4 bits (54), Expect(2) = 1e-12
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +2
Query: 3 GATIPIVEGVEKLGNNI 19
G PIVEGVEK NNI
Sbjct: 74 GQQSPIVEGVEKSRNNI 124
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 70.9 bits (172), Expect = 2e-12
Identities = 48/183 (26%), Positives = 89/183 (48%), Gaps = 2/183 (1%)
Frame = +1
Query: 312 PKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLY-QKSSKFCSDFEACID 370
P+TLLTD + + +AI+V MP++ H C W I L + ++ +DF +
Sbjct: 4 PQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFHRLYN 183
Query: 371 LHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFN 430
L E +F SW ++ ++ + + + ++ L+ WA ++R++F AG+ S +ES N
Sbjct: 184 LEMXE-DFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESIN 360
Query: 431 AELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKR-APMLVQAGNI 489
++ +L + +F IV N+ + A + K+ K +K +P+ A I
Sbjct: 361 VFIQRFLSAQSQPXRFLQQVADIVD--FNDRAGAKQXMQRKMQKXCLKTGSPIESHAATI 534
Query: 490 YTP 492
TP
Sbjct: 535 LTP 543
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 68.2 bits (165), Expect = 1e-11
Identities = 41/110 (37%), Positives = 59/110 (53%), Gaps = 3/110 (2%)
Frame = +3
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEG---KRGV 92
P + M+F S A FY+ Y RRIGF +R + +S+ + L + F C KEG K+ V
Sbjct: 315 PFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRVNNELIGQDFVCSKEGFRAKKYV 494
Query: 93 DKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPG 142
++D + TR GC A + ++L R GK+ V FV +H H L PG
Sbjct: 495 HRKDRVLPPPPA-TREGCQAMIRLAL-RDEGKWVVTKFVKEHTHKLMSPG 638
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 55.5 bits (132), Expect(2) = 6e-11
Identities = 31/74 (41%), Positives = 44/74 (58%), Gaps = 2/74 (2%)
Frame = +2
Query: 69 NKSKKDGILTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKI--GKYK 126
++S+KDG R C KEG R DKR+ + ++ R ETR GC A +++ RKI GK+
Sbjct: 137 SRSRKDGTAIGRALVCNKEGYRMPDKREKIVRQ-RAETRVGCRAMIMM---RKINSGKWV 304
Query: 127 VVDFVAQHNHLLEP 140
+ FV +H H L P
Sbjct: 305 ITKFVKEHTHPLNP 346
Score = 30.0 bits (66), Expect(2) = 6e-11
Identities = 13/26 (50%), Positives = 17/26 (65%)
Frame = +3
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGF 61
P++ EFESEAAA+ FYN G+
Sbjct: 33 PYVGQEFESEAAAHAFYNSIXAARGW 110
>TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43280
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1177
Score = 60.1 bits (144), Expect = 3e-09
Identities = 36/96 (37%), Positives = 54/96 (55%), Gaps = 1/96 (1%)
Frame = +1
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGK-RGVDK 94
P+ MEF SE AA FY++Y+R +GF +R +S +DG + +RRF C KEG +
Sbjct: 301 PYEGMEFVSEDAAKIFYDEYARHVGFVMRVMSCRRSXRDGRILARRFGCNKEGHCVSIQG 480
Query: 95 RDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDF 130
+ ++ R TR GC A + I D+ GK+ + F
Sbjct: 481 KLGPVRKPRASTREGCKAMIHIKYDQS-GKWMITXF 585
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 59.7 bits (143), Expect = 4e-09
Identities = 26/80 (32%), Positives = 42/80 (52%)
Frame = +1
Query: 304 LKAMGGEKPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCS 363
L AM G P ++ TD D + AI V P T H C W I + Q ++H++ + F +
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 364 DFEACIDLHEEEGEFLNSWN 383
+F C++L + EF + W+
Sbjct: 181 EFHKCVNLTDSIDEFESCWS 240
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 57.8 bits (138), Expect = 1e-08
Identities = 27/56 (48%), Positives = 34/56 (60%)
Frame = +1
Query: 33 DYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEG 88
D P MEFESE AA FYN Y+RR+GF R +S++DG + R+ C KEG
Sbjct: 184 DLEPSEGMEFESEEAAKAFYNSYARRVGFSTRVSSSRRSRRDGAIIQRQXVCAKEG 351
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 54.3 bits (129), Expect = 2e-07
Identities = 26/73 (35%), Positives = 42/73 (56%), Gaps = 1/73 (1%)
Frame = -3
Query: 513 LKEGLYVVTNYDN-NKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQ 571
L G Y++ + + ER V+ +++ C C++FE+ GILC HAL++L N +P+
Sbjct: 792 LYNGSYIIRHSKKLDGERHVLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLPE 613
Query: 572 HYILKRWTRDARL 584
Y L RW R+ L
Sbjct: 612 KYYLSRWHRERSL 574
>TC92547 similar to PIR|A56235|A56235 transcription activator MafB - chicken,
partial (6%)
Length = 1158
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/82 (40%), Positives = 45/82 (54%), Gaps = 5/82 (6%)
Frame = +3
Query: 38 LEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKRDH 97
L++EF AYEFY +Y + GF IR+ + GI+T R F C K G R DK+ H
Sbjct: 924 LKLEFGPADEAYEFYYRYGKCKGFSIRKG-DVRRNSSGIITMREFVCNKNGLR--DKK-H 1091
Query: 98 LTKEGRVE-----TRTGCDARM 114
L++ R TRT C+AR+
Sbjct: 1092 LSRNDRKRDHRRLTRTNCEARL 1157
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,287,111
Number of Sequences: 36976
Number of extensions: 304795
Number of successful extensions: 1401
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1380
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1387
length of query: 692
length of database: 9,014,727
effective HSP length: 103
effective length of query: 589
effective length of database: 5,206,199
effective search space: 3066451211
effective search space used: 3066451211
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126006.3


