

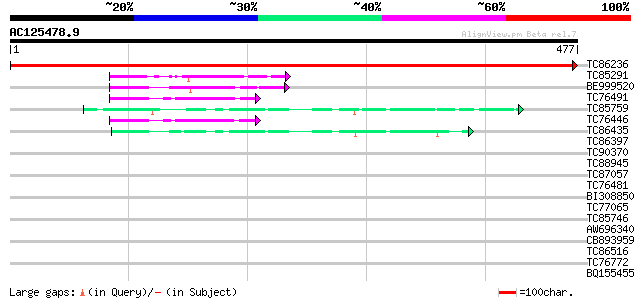
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.9 + phase: 0
(477 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86236 similar to PIR|T05311|T05311 sulfolipid biosynthesis pro... 971 0.0
TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (E... 51 9e-07
BE999520 homologue to SP|O65781|GAE2 UDP-glucose 4-epimerase GEP... 50 2e-06
TC76491 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase ... 49 3e-06
TC85759 similar to PIR|T48072|T48072 dTDP-glucose 4-6-dehydratas... 44 1e-04
TC76446 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase ... 43 3e-04
TC86435 homologue to PIR|T51252|T51252 dTDPglucose 4 6-dehydrata... 42 7e-04
TC86397 homologue to PIR|T51252|T51252 dTDPglucose 4 6-dehydrata... 40 0.002
TC90370 similar to GP|17473549|gb|AAL38251.1 dTDP-glucose 4-6-de... 40 0.002
TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.... 38 0.010
TC87057 similar to PIR|T48135|T48135 nucleotide sugar epimerase-... 35 0.063
TC76481 similar to PIR|G84677|G84677 probable dTDP-glucose 4-6-d... 34 0.11
BI308850 homologue to GP|13591616|db UDP-D-glucuronate carboxy-l... 34 0.14
TC77065 similar to PIR|A85356|A85356 nucleotide sugar epimerase-... 32 0.70
TC85746 similar to GP|16226201|gb|AAL16101.1 AT3g63140/T20O10_24... 32 0.70
AW696340 similar to SP|P39869|NIA_ Nitrate reductase (EC 1.6.6.1... 29 4.5
CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic ... 28 5.9
TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_... 28 5.9
TC76772 homologue to SP|O82043|ILV5_PEA Ketol-acid reductoisomer... 28 5.9
BQ155455 similar to GP|21553697|gb| unknown {Arabidopsis thalian... 28 7.7
>TC86236 similar to PIR|T05311|T05311 sulfolipid biosynthesis protein SQD1
chloroplast [validated] - Arabidopsis thaliana, partial
(83%)
Length = 1918
Score = 971 bits (2510), Expect = 0.0
Identities = 477/477 (100%), Positives = 477/477 (100%)
Frame = +1
Query: 1 MAQLLSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAV 60
MAQLLSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAV
Sbjct: 160 MAQLLSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAV 339
Query: 61 VNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRR 120
VNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRR
Sbjct: 340 VNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRR 519
Query: 121 LFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHF 180
LFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHF
Sbjct: 520 LFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHF 699
Query: 181 GEQRSAPYSMIDRSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIE 240
GEQRSAPYSMIDRSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIE
Sbjct: 700 GEQRSAPYSMIDRSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIE 879
Query: 241 EGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRT 300
EGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRT
Sbjct: 880 EGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRT 1059
Query: 301 DETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVEL 360
DETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVEL
Sbjct: 1060DETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVEL 1239
Query: 361 AIANPANPGEFRVFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVELEEHYYNC 420
AIANPANPGEFRVFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVELEEHYYNC
Sbjct: 1240AIANPANPGEFRVFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVELEEHYYNC 1419
Query: 421 KNTKLVDLGLKPHFLSDSLIDSLLNFAVQYKDRVDTKQIMPGVSWRKVGVKTKTLTS 477
KNTKLVDLGLKPHFLSDSLIDSLLNFAVQYKDRVDTKQIMPGVSWRKVGVKTKTLTS
Sbjct: 1420KNTKLVDLGLKPHFLSDSLIDSLLNFAVQYKDRVDTKQIMPGVSWRKVGVKTKTLTS 1590
>TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP- galactose 4-epimerase). [Garden
pea], partial (78%)
Length = 1018
Score = 51.2 bits (121), Expect = 9e-07
Identities = 43/157 (27%), Positives = 81/157 (51%), Gaps = 5/157 (3%)
Frame = +1
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
++++V GG G+ G T L L G+ V+I+DN FD+ + I+++ DR+
Sbjct: 202 QKILVTGGSGFIGTHTVLQLLQGGFAVSIIDN-----FDNSV-------IAAV-DRV--- 333
Query: 145 KSLTG----KSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQ 200
+ L G ++++ +GD+ + L + F + DAV+HF ++ S+ + R
Sbjct: 334 RELVGPQLSQNLDFTLGDLRIKDDLEKLFSKTKFDAVIHFAGLKAVGESVANPRRYF--- 504
Query: 201 QNNVVGTLNVLFAIKEYREDC-HLVKLGTMGEYGTPN 236
NN+VGT+N+ + +Y +C +V + YG P+
Sbjct: 505 DNNLVGTINLYEVMAKY--NCKKMVFSSSATVYGQPD 609
>BE999520 homologue to SP|O65781|GAE2 UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (46%)
Length = 499
Score = 50.1 bits (118), Expect = 2e-06
Identities = 45/154 (29%), Positives = 69/154 (44%), Gaps = 3/154 (1%)
Frame = +3
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K ++V GG GY G T L L GY+V +VDNL +S Q I
Sbjct: 3 KTILVTGGAGYIGSHTVLQLLLGGYKVVVVDNL----------------DNSSQKSIDRV 134
Query: 145 KSLTGK---SIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQ 201
K L G ++ + D+ + + L + F S + DAV+HF ++ S+ + +
Sbjct: 135 KKLAGDFAGNLSFHKLDLRDRDGLEKIFSSTKFDAVIHFAGLKAVGESV---QKPLLYYD 305
Query: 202 NNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTP 235
NN++GT+ VLF + LV + YG P
Sbjct: 306 NNLIGTI-VLFEVMAAHGCKKLVFSSSATVYGWP 404
>TC76491 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (96%)
Length = 1805
Score = 49.3 bits (116), Expect = 3e-06
Identities = 36/127 (28%), Positives = 63/127 (49%)
Frame = +1
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K V+V GG GY G T L L G++ IVDNL D+ + ++ R++
Sbjct: 271 KTVLVTGGAGYIGSHTVLQLLLGGFKTVIVDNL-----------DNSSEVAV--RRVKEL 411
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
+ GK++ + D+ + L + F S + DAV+HF ++ S+ + + NN+
Sbjct: 412 AAEFGKNLNFHKVDLRDKAALEQIFSSTKFDAVIHFAGMKAVGESV---QKPLLYYNNNL 582
Query: 205 VGTLNVL 211
+GT+ +L
Sbjct: 583 IGTITLL 603
>TC85759 similar to PIR|T48072|T48072 dTDP-glucose 4-6-dehydratase homolog
D18 - Arabidopsis thaliana, partial (84%)
Length = 1796
Score = 44.3 bits (103), Expect = 1e-04
Identities = 90/379 (23%), Positives = 141/379 (36%), Gaps = 9/379 (2%)
Frame = +3
Query: 63 ASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLV---R 119
AS + + + P+ G K R++V GG G+ G L +G V +VDN +
Sbjct: 462 ASVVHSSGKIPL----GIKRKGLRIVVTGGAGFVGSHLVDRLMARGDSVIVVDNFFTGRK 629
Query: 120 RLFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVH 179
H G EL D+ E L E D + H
Sbjct: 630 ENVMHHFG---------------------NPRFELIRHDVVEPLLL-------EVDQIYH 725
Query: 180 FGEQRSAPYSMID-RSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNID 238
+ P S + + V T + NVVGTLN+L K R + T YG P
Sbjct: 726 L----ACPASPVHYKFNPVKTIKTNVVGTLNMLGLAK--RVGARFLLTSTSEVYGDP--- 878
Query: 239 IEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRAT-----DLNQG 293
L +P++ + + +++ + +C G R D ++G
Sbjct: 879 ----------------LEHPQKETYWGNVNPIG-----VRSCYDEGKRTAETLTMDYHRG 995
Query: 294 VVYGVRTDETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRD 353
GV + R D G ++ F QA PLTVYG G QTR+F + D
Sbjct: 996 A--GVEVRIARIFNTYGPRMCLDD--GRVVSNFVAQALRKEPLTVYGDGKQTRSFQYVSD 1163
Query: 354 TVQCVELAIANPANPGEFRVFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVEL 413
V+ + + + + G F + N +F + ELA++V E + D K + N +
Sbjct: 1164LVEGL-MRLMEGEHVGPFNLGN--PGEFTMLELAKVVQ---ETIDPDAKIVYRDNTEDDP 1325
Query: 414 EEHYYNCKNTKLVDLGLKP 432
+ + N K LG +P
Sbjct: 1326HKRKPDISNAK-EHLGWEP 1379
>TC76446 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (55%)
Length = 703
Score = 42.7 bits (99), Expect = 3e-04
Identities = 34/127 (26%), Positives = 59/127 (45%)
Frame = +1
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K V+V GG GY G T L L G++ +VDNL D+ + ++ R++
Sbjct: 52 KTVLVTGGAGYIGSHTVLQLLLGGFKSIVVDNL-----------DNSSEVAI--HRVKEL 192
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
G ++ + D+ + L + F S DAV+HF ++ S + + NN+
Sbjct: 193 AGEFGNNLSFHKVDLRDRAALEQIFGSTTFDAVIHFAGLKAVGES---AQKPLLYYNNNL 363
Query: 205 VGTLNVL 211
+GT+ +L
Sbjct: 364 IGTITLL 384
>TC86435 homologue to PIR|T51252|T51252 dTDPglucose 4 6-dehydratase (EC
4.2.1.46) [imported] - chickpea, complete
Length = 1730
Score = 41.6 bits (96), Expect = 7e-04
Identities = 75/317 (23%), Positives = 116/317 (35%), Gaps = 12/317 (3%)
Frame = +3
Query: 86 RVMVIGGDGYCGWATALHL-SNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
R++V GG G+ G L N+ EV + DN + +D ++ W
Sbjct: 432 RILVTGGAGFIGSHLVDRLMENEKNEVIVADNF----------------FTGSKDNLKKW 563
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQNN 203
+ EL D+ E + E D + H + P S I + V T + N
Sbjct: 564 --IGHPRFELIRHDVTETLMI-------EVDQIYHL----ACPASPIFYKYNPVKTIKTN 704
Query: 204 VVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASS 263
V+GTLN+L K R ++ T YG P L +P+ S
Sbjct: 705 VIGTLNMLGLAK--RVGARILLTSTSEVYGDP-------------------LEHPQPESY 821
Query: 264 FYHLSKVHDSHNIAFTCKAWGIRATD---LNQGVVYGVRTDETAMHEELCNRFDYDAIFG 320
+ +++ + +C G R + + +G+ + R + D G
Sbjct: 822 WGNVNPIG-----VRSCYDEGKRVAETLMFDYHRQHGIEIRVARIFNTYGPRMNIDD--G 980
Query: 321 TALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEFRV 373
++ F QA G LTV G QTR+F + D V + + N NPGE
Sbjct: 981 RVVSNFIAQALRGESLTVQAPGTQTRSFCYVSDLVDGLIRLMGGSDTGPINLGNPGE--- 1151
Query: 374 FNQFTEQFKVTELAELV 390
F +TELAE V
Sbjct: 1152-------FTMTELAETV 1181
>TC86397 homologue to PIR|T51252|T51252 dTDPglucose 4 6-dehydratase (EC
4.2.1.46) [imported] - chickpea, complete
Length = 1946
Score = 40.4 bits (93), Expect = 0.002
Identities = 77/307 (25%), Positives = 116/307 (37%), Gaps = 2/307 (0%)
Frame = +1
Query: 86 RVMVIGGDGYCGWATALHL-SNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
R++V GG G+ G L N+ EV + DN + +D ++ W
Sbjct: 289 RILVTGGAGFIGSHLVDRLMENEKNEVIVADNY----------------FTGCKDNLKKW 420
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQNN 203
+ EL D+ E + E D + H + P S I + V T + N
Sbjct: 421 --IGHPRFELIRHDVTETLLV-------EVDRIYHL----ACPASPIFYKYNPVKTIKTN 561
Query: 204 VVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASS 263
V+GTLN+L K R ++ T YG P I + T+ G + P S
Sbjct: 562 VIGTLNMLGLAK--RVGARILLTSTSEVYGDPLIHPQPE----TYWGNVN----PIGVRS 711
Query: 264 FYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRFDYDAIFGTAL 323
Y K +A T + +G+ + R + D G +
Sbjct: 712 CYDEGK-----RVAETLM--------FDYHRQHGLEIRIARIFNTYGPRMNIDD--GRVV 846
Query: 324 NRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTEQFKV 383
+ F QA G PLTV G QTR+F + D V + + + N G + N +F +
Sbjct: 847 SNFIAQAIRGEPLTVQLPGTQTRSFCYVSDMVDGL-IRLMEGENTGPINIGN--PGEFTM 1017
Query: 384 TELAELV 390
TELAE V
Sbjct: 1018TELAENV 1038
>TC90370 similar to GP|17473549|gb|AAL38251.1 dTDP-glucose
4-6-dehydratase-like protein {Arabidopsis thaliana},
partial (31%)
Length = 665
Score = 40.0 bits (92), Expect = 0.002
Identities = 26/71 (36%), Positives = 39/71 (54%)
Frame = +2
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTE 379
G ++ F QA PLTVYG G QTR+F + D V + A+ + + G F + N
Sbjct: 8 GRVVSNFVAQAIRKQPLTVYGDGKQTRSFQYVSDLVNGL-AALMDGEHVGPFNLGN--PG 178
Query: 380 QFKVTELAELV 390
+F + ELA++V
Sbjct: 179 EFTMLELAQVV 211
>TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.120 -
Arabidopsis thaliana, partial (94%)
Length = 1345
Score = 37.7 bits (86), Expect = 0.010
Identities = 84/383 (21%), Positives = 132/383 (33%), Gaps = 23/383 (6%)
Frame = +3
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K+V+V G GY G L +GY V ++ + P S++
Sbjct: 105 KKVVVTGASGYLGGKLCNSLHRQGYSVKVI----------------VRPTSNL------- 215
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
S S E+ GDI +F L F D V F + D S+ + NV
Sbjct: 216 -SALPPSTEIVYGDITDFSSLLSAFS----DCSVVFHLAALVEPWLPDPSKFITV---NV 371
Query: 205 VGTLNVLFAIKEYREDCHLVKLGTMGEYG-TPNIDIEEGYITITHNGRTDTLPYPKQASS 263
G NVL A+K+ + LV + G T +E + H+ R Y K
Sbjct: 372 EGLKNVLEAVKQTKTVEKLVYTSSFFALGPTDGAIADENQV---HHERFFCTEYEK---- 530
Query: 264 FYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRFDYDAIFGTAL 323
SKV + IA + G+ L GV+YG G +
Sbjct: 531 ----SKV-ATDKIALQAASEGVPIVLLYPGVIYG----------------PGKVTAGNVV 647
Query: 324 NRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTEQF-K 382
+ V+ G GKG +F + D V+ +A GE + F +
Sbjct: 648 AKMLVERFSGRLPGYIGKGNDKFSFSHVDDVVE-GHIAAMKKGQIGERYLLTGENASFNQ 824
Query: 383 VTELAELVTKAGE---------------------KLGLDVKTISVPNPRVELEEHYYNCK 421
V ++A ++T + ++ + IS P V Y+C+
Sbjct: 825 VFDMAAVITNTSKPMVSIPLCVIEAYGWLLVLISRITGKLPFISPPTVHVLRHRWEYSCE 1004
Query: 422 NTKLVDLGLKPHFLSDSLIDSLL 444
K+ +L KP L + L + L+
Sbjct: 1005KAKM-ELDYKPRSLREGLAEVLI 1070
>TC87057 similar to PIR|T48135|T48135 nucleotide sugar epimerase-like
protein - Arabidopsis thaliana, partial (80%)
Length = 1655
Score = 35.0 bits (79), Expect = 0.063
Identities = 52/214 (24%), Positives = 78/214 (36%), Gaps = 1/214 (0%)
Frame = +2
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K V+V G G+ G AL L +G D LG+D+ + +
Sbjct: 314 KIVLVTGSAGFVGSHVALALKRRG--------------DGVLGIDNFNRYYDVNLKHARQ 451
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
K L I + GDI + L + F V+H Q Y+M + + V+ +N+
Sbjct: 452 KLLERAGIFVVEGDINDGTLLKKLFDVVPFTHVMHLAAQAGVRYAMQNPNSYVH---SNL 622
Query: 205 VGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASSF 264
G +L A K +V + YG + + RTD Q +S
Sbjct: 623 AGFTVLLEACKSANPQPAIVWASSSSVYGL------NSKVPFSEKDRTD------QPASL 766
Query: 265 YHLSKVHDSHNIAFTCK-AWGIRATDLNQGVVYG 297
Y +K IA T +G+ T L VYG
Sbjct: 767 YAATK-KAGEGIAHTYNHIYGLSITALRFFTVYG 865
>TC76481 similar to PIR|G84677|G84677 probable dTDP-glucose 4-6-dehydratase
[imported] - Arabidopsis thaliana, partial (98%)
Length = 1811
Score = 34.3 bits (77), Expect = 0.11
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 5/101 (4%)
Frame = +3
Query: 333 GHPLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELV 390
G PL + G R F+ I+D ++ V L I NP AN F V N + V +LAE++
Sbjct: 888 GEPLKLVDGGESQRTFVYIKDAIEAVLLMIENPARANGHIFNVGNP-NNEVTVRQLAEMM 1064
Query: 391 TKAGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVDL 428
+ K+ + P V +E Y Y+ + ++ D+
Sbjct: 1065IQVYSKVS-GTQPPETPTIDVSSKEFYGEGYDDSDKRIPDM 1184
>BI308850 homologue to GP|13591616|db UDP-D-glucuronate carboxy-lyase {Pisum
sativum}, partial (60%)
Length = 629
Score = 33.9 bits (76), Expect = 0.14
Identities = 26/78 (33%), Positives = 34/78 (43%), Gaps = 7/78 (8%)
Frame = +1
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEFR 372
G ++ F QA G LTV G QTR+F + D V + + N +PGE
Sbjct: 394 GRVVSNFIAQALRGESLTVQAPGTQTRSFCYVSDLVDGLIRLMGGSDTGPINLGHPGE-- 567
Query: 373 VFNQFTEQFKVTELAELV 390
F +TELAE V
Sbjct: 568 --------FTMTELAETV 597
>TC77065 similar to PIR|A85356|A85356 nucleotide sugar epimerase-like
protein [imported] - Arabidopsis thaliana, partial (97%)
Length = 1679
Score = 31.6 bits (70), Expect = 0.70
Identities = 44/211 (20%), Positives = 75/211 (34%)
Frame = +2
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V G G+ G +L L +G D +GLD+ +
Sbjct: 371 VLVTGAAGFIGSHVSLALKRRG--------------DGVVGLDNFNDYYDPSLKKARKAL 508
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
L + + + GD+ + + L++ F V+H Q Y+M + V +N+ G
Sbjct: 509 LQSRGVFIVHGDLNDAKLLAKLFDVVAFTHVMHLAAQAGVRYAMENPMSYV---NSNIAG 679
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASSFYH 266
+ +L A K +V + YG + + + RTD Q +S Y
Sbjct: 680 LVTLLEACKTANPQPSIVWASSSSVYGL------NEKVPFSESDRTD------QPASLYA 823
Query: 267 LSKVHDSHNIAFTCKAWGIRATDLNQGVVYG 297
+K +G+ T L VYG
Sbjct: 824 ATKKAGEEITHTYNHIYGLSITGLRFFTVYG 916
>TC85746 similar to GP|16226201|gb|AAL16101.1 AT3g63140/T20O10_240
{Arabidopsis thaliana}, partial (80%)
Length = 1347
Score = 31.6 bits (70), Expect = 0.70
Identities = 30/118 (25%), Positives = 49/118 (41%), Gaps = 4/118 (3%)
Frame = +1
Query: 1 MAQLLSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAV 60
MA L SSS +L F++++ L H C+ S + ++ FL S +
Sbjct: 64 MAMLASSSPTLLFTSTSSNLLPLSHSCTLRLSFSSSLQSTSLSISSTFL-SHPSLTSKRL 240
Query: 61 VNASTISTGQEAPVQTSSGDPFKPKRVMVI----GGDGYCGWATALHLSNKGYEVAIV 114
N +T+S A + K+V++I GG G+ A L G+ V I+
Sbjct: 241 ANHATLSISASAA---------EKKKVLIINTNSGGHAVIGFYFAKELLGAGHSVTIL 387
>AW696340 similar to SP|P39869|NIA_ Nitrate reductase (EC 1.6.6.1) (NR).
{Lotus japonicus}, partial (12%)
Length = 336
Score = 28.9 bits (63), Expect = 4.5
Identities = 20/55 (36%), Positives = 28/55 (50%)
Frame = -1
Query: 10 SLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAVVNAS 64
S+TF +SN LKPFH ST + P R+ +R Q SLA+++ S
Sbjct: 333 SITFISSNNNPLKPFHHPSTDKT------RYNNPHRKPMIRSQ----SLAILHVS 199
>CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (28%)
Length = 791
Score = 28.5 bits (62), Expect = 5.9
Identities = 13/58 (22%), Positives = 30/58 (51%)
Frame = -2
Query: 6 SSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAVVNA 63
++SC T + + +KPF S++ ++ C +S T ++ F P+ S ++ ++
Sbjct: 625 TASCVNTSNCGHHVTIKPFFDVSSNPFGSLTCSDSLTTHKKYFSLSFNPKASSSICSS 452
>TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_190
{Arabidopsis thaliana}, partial (83%)
Length = 1499
Score = 28.5 bits (62), Expect = 5.9
Identities = 12/45 (26%), Positives = 21/45 (46%)
Frame = +2
Query: 67 STGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEV 111
++ E V G P +P ++V+G G G ++GY+V
Sbjct: 272 NSNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDV 406
>TC76772 homologue to SP|O82043|ILV5_PEA Ketol-acid reductoisomerase
chloroplast precursor (EC 1.1.1.86), complete
Length = 1996
Score = 28.5 bits (62), Expect = 5.9
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 8/99 (8%)
Frame = +3
Query: 5 LSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKSLAV---- 60
++SSCS SAS+K KP +TSF+ + F +L+ + K R+ +AV
Sbjct: 45 VTSSCSTAISASSKTLTKP---TTTSFA------AANLSFSKLYPQSVKARRCIAVGGAV 197
Query: 61 ----VNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGY 95
V+A + + +TS FK +RV + G + +
Sbjct: 198 GARMVSAPPATLPAKLDFETSI---FKKERVNLAGHEEF 305
>BQ155455 similar to GP|21553697|gb| unknown {Arabidopsis thaliana}, partial
(29%)
Length = 655
Score = 28.1 bits (61), Expect = 7.7
Identities = 17/33 (51%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Frame = +1
Query: 2 AQLLSSSCSLTFSASNKP-CLKPFHQCSTSFSN 33
+ L S SCS S+SN P LK F Q S+S SN
Sbjct: 157 SSLFSRSCSTRGSSSNSPLLLKSFSQKSSSTSN 255
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,842,436
Number of Sequences: 36976
Number of extensions: 201489
Number of successful extensions: 1021
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1008
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1019
length of query: 477
length of database: 9,014,727
effective HSP length: 100
effective length of query: 377
effective length of database: 5,317,127
effective search space: 2004556879
effective search space used: 2004556879
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC125478.9


