

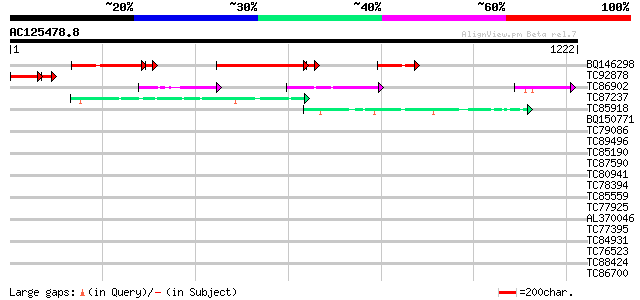
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.8 + phase: 0
(1222 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ146298 403 e-119
TC92878 homologue to PIR|F85062|F85062 hypothetical protein AT4g... 132 1e-44
TC86902 similar to GP|4585899|gb|AAD25561.1| unknown protein {Ar... 111 1e-24
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 44 3e-04
TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreu... 43 9e-04
BQ150771 GP|4028979|emb glutamate permease {synthetic construct}... 41 0.003
TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein l... 40 0.007
TC89496 similar to GP|4585899|gb|AAD25561.1| unknown protein {Ar... 39 0.016
TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 36 0.10
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 35 0.23
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 33 0.88
TC78394 similar to GP|10177020|dbj|BAB10258. leucine zipper prot... 32 1.2
TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC ... 32 1.2
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 32 1.2
AL370046 GP|10177686|db emb|CAB62594.1~gene_id:MUL8.6~similar to... 32 1.5
TC77395 similar to GP|4874305|gb|AAD31367.1| expressed protein {... 32 1.5
TC84931 SP|P75780|YBIL_ECOLI Probable tonB-dependent receptor yb... 32 2.0
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 32 2.0
TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Ar... 32 2.0
TC86700 similar to GP|18377835|gb|AAL67104.1 At1g24120/F3I6_4 {A... 31 3.3
>BQ146298
Length = 691
Score = 403 bits (1035), Expect(2) = e-119
Identities = 198/199 (99%), Positives = 199/199 (99%)
Frame = +3
Query: 446 LLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARN 505
LLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARN
Sbjct: 3 LLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARN 182
Query: 506 DKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGT 565
DKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGT
Sbjct: 183 DKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGT 362
Query: 566 STTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGN 625
STTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGN
Sbjct: 363 STTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGN 542
Query: 626 NSSCHNYNETDSDMDEEKK 644
NSSCHNYNETDSDMDEEK+
Sbjct: 543 NSSCHNYNETDSDMDEEKE 599
Score = 125 bits (315), Expect(2) = 1e-35
Identities = 77/172 (44%), Positives = 106/172 (60%), Gaps = 10/172 (5%)
Frame = +3
Query: 134 QKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNS--QQNVKNVSMESSPFKPHDAPPSTVN 191
Q SK+ N R+ + T+ ++ ++N ++ +++S+ K P
Sbjct: 99 QSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPV---- 266
Query: 192 ECDTSTKDKHMVTD--YEVLQKSSTQEEPKPG-STTSVAYGVQERDQKYIKKWHLMYKHA 248
CDT ++ + + Y+V +KS +E+ K G STT V YGVQERDQKYIKKWHLMYK A
Sbjct: 267 -CDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQA 443
Query: 249 VLSNTGKCDNKVPLVEKEKEGGEED----NEGNN-SYRNYSETDSDMDDEKK 295
VLSNTGK DNK+P+V K+KEG E+ N GNN S NY+ETDSDMD+EK+
Sbjct: 444 VLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKE 599
Score = 63.5 bits (153), Expect = 5e-10
Identities = 38/89 (42%), Positives = 55/89 (61%)
Frame = +3
Query: 794 LLIEAFETLRPIQDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEF 853
LL+EAFET+RP+QDAENG ++SATVES N +QSLDASS S + + + F E
Sbjct: 3 LLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDF------EV 164
Query: 854 SDKASDNPMPELCKPIKPVETISSCHEEA 882
+++A ++ + CK T+ S +A
Sbjct: 165 TERARNDKNMDACKKNDESATVKSTATKA 251
Score = 45.8 bits (107), Expect(2) = e-119
Identities = 22/26 (84%), Positives = 25/26 (95%)
Frame = +1
Query: 641 EEKKNVIELVQKAFDEILLPETEDLS 666
++KKNVIELV KAFDEILLPETE+LS
Sbjct: 589 KKKKNVIELVXKAFDEILLPETENLS 666
Score = 43.5 bits (101), Expect(2) = 1e-35
Identities = 21/25 (84%), Positives = 23/25 (92%)
Frame = +1
Query: 293 EKKNVIELVQKAFDEILLPEVEDLS 317
+KKNVIELV KAFDEILLPE E+LS
Sbjct: 592 KKKNVIELVXKAFDEILLPETENLS 666
>TC92878 homologue to PIR|F85062|F85062 hypothetical protein AT4g04980
[imported] - Arabidopsis thaliana, partial (1%)
Length = 608
Score = 132 bits (333), Expect(2) = 1e-44
Identities = 67/69 (97%), Positives = 68/69 (98%)
Frame = +2
Query: 1 MKSSRSIKLLTVKGPKSTTKLYSESTDGIDGNNRNSTSDAGNKSQRVMTRRLSLKPVRIS 60
MKSSRSIKLLTVKGPKSTTKLYSESTDGIDGNNRNSTSDAGNKSQRVMTRRLSLKPVRIS
Sbjct: 302 MKSSRSIKLLTVKGPKSTTKLYSESTDGIDGNNRNSTSDAGNKSQRVMTRRLSLKPVRIS 481
Query: 61 AKKPSLHKA 69
KKPSLHK+
Sbjct: 482 XKKPSLHKS 508
Score = 67.0 bits (162), Expect(2) = 1e-44
Identities = 31/34 (91%), Positives = 32/34 (93%)
Frame = +3
Query: 68 KATCSSTIKDSHFPNHIDLPQEGSSSQGVSAVKV 101
KATCSSTIKDSHFPNHIDLPQEGSSSQGVS +V
Sbjct: 504 KATCSSTIKDSHFPNHIDLPQEGSSSQGVSLXRV 605
>TC86902 similar to GP|4585899|gb|AAD25561.1| unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 1623
Score = 111 bits (278), Expect = 1e-24
Identities = 66/147 (44%), Positives = 89/147 (59%), Gaps = 17/147 (11%)
Frame = +1
Query: 1089 EGLNQKEQKMESGNGIVEERKE-------ESVSKEVN---KPNQKLS-------RNWSNL 1131
E +++ Q+ E N +EE+ E E K++ K N++ S N
Sbjct: 673 ENVDETSQENEDDNINIEEKDEIQLFDVLEGSIKDIQDQCKGNKRASCIIDEDEDTRGNR 852
Query: 1132 KKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYAL 1191
K V+ +R + +++R FNPREP +LPL P+ E EKV LRHQ M ERK E+WM+D AL
Sbjct: 853 KGVIRRKRNDEDDDELRNFNPREPNFLPLVPEKEKEKVDLRHQMMDERKNAEDWMVDCAL 1032
Query: 1192 RQVVSKLTPARKRKVELLVEAFETVVP 1218
RQ V+KL PARK+KV LLVEAFETV+P
Sbjct: 1033 RQAVNKLAPARKKKVALLVEAFETVIP 1113
Score = 86.7 bits (213), Expect = 5e-17
Identities = 60/214 (28%), Positives = 107/214 (49%), Gaps = 5/214 (2%)
Frame = +1
Query: 596 NTGKYDNKLPVVGKDKEGREQGDAVFNG--GNNSSC---HNYNETDSDMDEEKKNVIELV 650
NT + + + +D+ + + VF G G+N + ++Y++ D + + +
Sbjct: 481 NTQRSQIRRNIYLEDQPHNDMINFVFEGKIGSNETQEIGYSYDDIGCDQSSLANEIFDYL 660
Query: 651 QKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENK 710
A + + ET + DD +L + EG +++ + + +
Sbjct: 661 TNAEENV--DETSQENEDDNINIEEKDEIQLFDVLEGSIKDIQ-DQCKGNKRASCIIDED 831
Query: 711 PKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEE 770
+ + K +I KR + D++RN NPR P LP E EKV L Q +ERK +E+
Sbjct: 832 EDTRGNRKGVIRRKRNDEDDDELRNFNPREPNFLPLVPEKEKEKVDLRHQMMDERKNAED 1011
Query: 771 WMLDYALQKVISKLAPAQRQRVTLLIEAFETLRP 804
WM+D AL++ ++KLAPA++++V LL+EAFET+ P
Sbjct: 1012WMVDCALRQAVNKLAPARKKKVALLVEAFETVIP 1113
Score = 84.3 bits (207), Expect = 3e-16
Identities = 55/179 (30%), Positives = 92/179 (50%)
Frame = +1
Query: 278 NSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKS 337
N+ N ET + +D+ N+ E + ++L ++D+ + +GN+ +++
Sbjct: 664 NAEENVDETSQENEDDNINIEEKDEIQLFDVLEGSIKDIQDQ----CKGNKRASCIID-- 825
Query: 338 GGKIEERNTTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLP 397
E+ +T + K VI KR + +++RN N R P LP
Sbjct: 826 ----EDEDTR-------------------GNRKGVIRRKRNDEDDDELRNFNPREPNFLP 936
Query: 398 SDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRP 456
E EKV L Q +ERK +E+WM+D AL++ ++KLAPA++++V LLVEAFET+ P
Sbjct: 937 LVPEKEKEKVDLRHQMMDERKNAEDWMVDCALRQAVNKLAPARKKKVALLVEAFETVIP 1113
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 44.3 bits (103), Expect = 3e-04
Identities = 105/542 (19%), Positives = 195/542 (35%), Gaps = 27/542 (4%)
Frame = +1
Query: 132 KSQKSTKKDGRSKQVGNAR-----KGTQKTKTVHSEDGNSQQNV--KNVSMESSPFKPHD 184
K + KDG Q N K Q+ + E+ + Q+N KN E + +
Sbjct: 160 KQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITE 339
Query: 185 APPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLM 244
NE + T K + S +E+ G +E + +
Sbjct: 340 EKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGT---EGGEK 510
Query: 245 YKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKA 304
+H + DN+V EK E EE+ G+N+ + + +D++ + ++
Sbjct: 511 EEHDKIKEDTSSDNQVQDGEKNNEAREENYSGDNA------SSAVVDNKSQESSNKTEEQ 672
Query: 305 FDEILLPEVEDLSSEGHSKSRGNETDEVLLEKS-GGKIEERNTTTFTESPKEVPKMESKQ 363
FD+ E E L S+ +S TD + + S G + E+ T ++PK +Q
Sbjct: 673 FDKKEKNEFE-LESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQ 849
Query: 364 KSWSHLKKVILLKRFVKALE---KVRNINSRRPRQLPSDANFEAE-KVLLNRQTSEERKK 419
K K V+ + + N + + + DAN + E LN T+ +
Sbjct: 850 KQEQEQNNT--TKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALN--TTPNNED 1017
Query: 420 SEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQS 479
S+ + + + + T E +T + +G + + S +N +
Sbjct: 1018SKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDN 1197
Query: 480 LDASSN----------HSKEEINDRRDFEVTERARNDKNMDACKKN---DESATVKSTAT 526
DA+SN S+++ ++ E + + N ++ + N DESA ++
Sbjct: 1198KDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSNTTSDESANDNKDSS 1377
Query: 527 KAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKE-DLKHGTSTTDVPYGVQERDQKYIKKW 585
+ E +AEG + S + D K+ +T D G D +
Sbjct: 1378QVT---------TSSENSAEGNSNTENNSDENQNDSKNNENTND--SGNTSNDANVNENQ 1524
Query: 586 HLMYKQAVLS-NTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKK 644
+ Q S N G N+ V KE E + +NS+ ++T D+++
Sbjct: 1525NENAAQTKTSENEGDAQNE--SVESKKENNESAHKDVDNNSNSNDQGSSDTSVTQDDKES 1698
Query: 645 NV 646
V
Sbjct: 1699RV 1704
>TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreurys tristis},
partial (8%)
Length = 4117
Score = 42.7 bits (99), Expect = 9e-04
Identities = 98/523 (18%), Positives = 192/523 (35%), Gaps = 29/523 (5%)
Frame = -1
Query: 634 ETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSD-------DRSKSRSYGSDELLEKSE 686
ET +++E E Q ++I ET + SSD + ++ S+ EK E
Sbjct: 2818 ETSQQIEKELVETNEENQPKIEDIPEAETTEKSSDATETNQFEEKVKQAEISETSAEKEE 2639
Query: 687 GEREEMNATSFTETPKEAKK-TENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELP 745
E AT + KE +K ++ K + K + +++ V+A + E
Sbjct: 2638 KPELEPLATEEPKVTKEPEKESQEKSEEAEQPKAIAIVESTVEANE-----------EKT 2492
Query: 746 SDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISK---------LAPAQRQRVTLLI 796
+ A E K+ T E K E + + A +++ + + + V +
Sbjct: 2491 AAAISEETKIIEVEPT--ETVKEEPMVTEVATAEIVKEEPVVTEVEATETLKEELVATEV 2318
Query: 797 EAFETLRPIQDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDK 856
EA ET++ + A ++ + TV+ E + +D + + + L + E ++
Sbjct: 2317 EATETVKE-EPAATEVKVTETVKD-EPVVTEVDPTETVKEEPLATEA--------EATET 2168
Query: 857 ASDNPMPELCKPIKPVETISSCHEEAPTKRMVDEVPEDLVSDLNTKTKDVIGGHGE---- 912
+ P+ +P + V+ E T+ + DE V T ++ + E
Sbjct: 2167 VKEEPVATEVEPTETVKEEPVATEVEATETVKDEPAATEVEATETVKEEPVATETEATET 1988
Query: 913 --------QFSVTKSLILNGIVRSLRSNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEA 964
+ T+++ + + + V E P + EPT +KD + E +E
Sbjct: 1987 VKEEPVATEVEATETVKEESVATEVEATETVKEEPVTTVVEPTEMVKDEPAATEFEATET 1808
Query: 965 PTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREIS 1024
T+ + T +T+V K K +T +++
Sbjct: 1807 VKDE------------------TIVTEVDPTETVKEEAVATEVEITKTVKEPLVTEVDLT 1682
Query: 1025 VSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTG 1084
S + PVV + P R +V E +++A + V++ + + + +P T+
Sbjct: 1681 ESVKVKPVVTEVDP---REMVKEEPVATEVQATETVKE--EPVATEVEP---TETVKGEP 1526
Query: 1085 GIYSEGLNQKEQKMESGNGIVEERKEESVSKEVNKPNQKLSRN 1127
+ NQKE + S ER+EE + P Q N
Sbjct: 1525 VVTEVEENQKEPEQHS-----TERREEEQPNASSIPEQSTETN 1412
>BQ150771 GP|4028979|emb glutamate permease {synthetic construct}, partial
(3%)
Length = 1234
Score = 40.8 bits (94), Expect = 0.003
Identities = 34/175 (19%), Positives = 67/175 (37%), Gaps = 10/175 (5%)
Frame = +1
Query: 127 RRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAP 186
RR + K + + R K++ R + + D NS + + S S+P +
Sbjct: 448 RRPEAKRPERGNEMYRKKEMDRKRGVREAANESYR*DTNSNTHERTTSRSSTPRRRDART 627
Query: 187 PSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYK 246
P T E T + +H ++ + ++ QE +P + + + H Y+
Sbjct: 628 PQTRGETQTRNRRQHASATHQT*KPAAIQELHRPTEHAEMQGQTENERRSTTPTTHNEYR 807
Query: 247 HAV----------LSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMD 291
++ + D + E+ ++ GEE NE +R +SET + D
Sbjct: 808 ARADNDHQSKGERAGDSTRPDARAARRERAEQRGEETNEKERRHRRHSETVTGRD 972
>TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein ligase 1
putative {Plasmodium falciparum 3D7}, partial (0%)
Length = 1418
Score = 39.7 bits (91), Expect = 0.007
Identities = 56/250 (22%), Positives = 88/250 (34%), Gaps = 16/250 (6%)
Frame = +2
Query: 133 SQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVNE 192
S + KD Q + + + SE G+S + + S P PP N
Sbjct: 59 SATAAVKDDPPPQEDKIEQHDENDEETSSEAGSSSEGGEEEEESSEEEAPKTTPPPKTNP 238
Query: 193 CDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHAVLSN 252
+ S +D E +++ T E+ P T + ++ + I K +N
Sbjct: 239 KNNSVED-------EDEEETDTDEDEPPPKTKPIEQTLKSGTKPSIAPARSGTKRPAENN 397
Query: 253 TGKCDNKVPLVE-----KEKEGGEEDNEGNNS---YRNYSETDS--------DMDDEKKN 296
K NK E KEKE E+DN N R ++E D D + N
Sbjct: 398 DTKQSNKKKTTEEKNKKKEKEPEEDDNSNNKKASFQRVFTEDDEVAILQGLVDFTAKSGN 577
Query: 297 VIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEV 356
AF +I+ V + + + L K KI+ + T TF++ P E
Sbjct: 578 DPTKHLSAFYQIVKKSVHFKVTLDQLRDKVRR----LRLKYENKIKSKKTPTFSK-PVEE 742
Query: 357 PKMESKQKSW 366
E +K W
Sbjct: 743 TMFELSKKIW 772
>TC89496 similar to GP|4585899|gb|AAD25561.1| unknown protein {Arabidopsis
thaliana}, partial (7%)
Length = 811
Score = 38.5 bits (88), Expect = 0.016
Identities = 20/46 (43%), Positives = 29/46 (62%)
Frame = +1
Query: 46 RVMTRRLSLKPVRISAKKPSLHKATCSSTIKDSHFPNHIDLPQEGS 91
R +T+ S K R S + S +ATCSST+KDS FP+++ L G+
Sbjct: 670 RTLTKTTSFKASRTSCPRKST-RATCSSTLKDSSFPSYLMLNHGGT 804
>TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (50%)
Length = 1496
Score = 35.8 bits (81), Expect = 0.10
Identities = 58/261 (22%), Positives = 105/261 (40%), Gaps = 12/261 (4%)
Frame = +3
Query: 936 PEAPSNRLDEPTTDIKDVVEKDQLEK---SEAPTSAVVESKNQLEKQGSTGLWFTVFKHM 992
P+ P+ + TT + VVEK+Q + A T+A V + T K
Sbjct: 144 PQKPAEEV--ATTTSETVVEKEQQADGVVAAAVTAAAVTAA-------------TTDKEA 278
Query: 993 VSD----MTENNSKTSTDVADE-KDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDA 1047
V+D + + K + VAD+ D D + SVS++ V+ ++P + +D
Sbjct: 279 VADPPPAVADEAEKPAEVVADKVADETVVDESKVSQSVSFKEETNVVSELPDVQKKALD- 455
Query: 1048 EVELRQI--EAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIV 1105
EL+Q+ EA+ E P P P+ + + + E+ + +V
Sbjct: 456 --ELKQLIQEALNKHEFTAPPPAPVKAPEPEVAVKEE---------KKPEEDEKKTEEVV 602
Query: 1106 EERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSE 1165
EE+K+E+V +E K +++ + EP+ EP+ E
Sbjct: 603 EEKKDEAVVEE-------------------------KKVDEEKGSTSEEPKVETAEPEKE 707
Query: 1166 DEKVQLRHQDMAER--KGTEE 1184
++KV+ ++ E+ TEE
Sbjct: 708 EKKVEETVVEVVEKIAASTEE 770
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 34.7 bits (78), Expect = 0.23
Identities = 45/184 (24%), Positives = 83/184 (44%), Gaps = 22/184 (11%)
Frame = +2
Query: 346 TTTFTESPKEVPKMESKQKSWSH--LKKVILLKRFVKALEK--------------VRNIN 389
T+T T +P+ V K +Q+ + L+ V + V+A K V I+
Sbjct: 284 TSTTTTTPRSVLKHHQQQQHSDNKSLQTVPQTRLRVRASSKAKESPKTPPEIVNRVSTIS 463
Query: 390 SRRPRQLPSDA--NFEAEK-VLLNR---QTSEERKKSEEWMLDYALQKVISKLAPAQRQR 443
S R + +P D N +A++ + +N+ EE + S + + + KV+ +AP +R+R
Sbjct: 464 STRAKSVPPDMKNNSKAKRSIFMNKVVKSIEEEVESSHKGSKEGEVAKVVV-VAPPRRRR 640
Query: 444 VTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERA 503
+ E P D + + +E NLI+SL + N K+E+N + + +
Sbjct: 641 I-------EEDDP--DVKEKKELLEKLEVSENLIKSLQSEINALKDELNQVKGLNIDLES 793
Query: 504 RNDK 507
+N K
Sbjct: 794 QNIK 805
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 32.7 bits (73), Expect = 0.88
Identities = 29/101 (28%), Positives = 46/101 (44%), Gaps = 1/101 (0%)
Frame = +2
Query: 238 IKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGG-EEDNEGNNSYRNYSETDSDMDDEKKN 296
++KW LM + + + K+ EKE EG EE EGN+ + E + DD +K+
Sbjct: 59 MRKWMLMXEVKETTEDKEEKEKIE-AEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKS 235
Query: 297 VIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKS 337
E +K + E E +E + E DE ++KS
Sbjct: 236 EEETEEKEEGD----EKEKTEAETEEE---EEADEDTIDKS 337
>TC78394 similar to GP|10177020|dbj|BAB10258. leucine zipper protein
{Arabidopsis thaliana}, partial (83%)
Length = 2109
Score = 32.3 bits (72), Expect = 1.2
Identities = 17/46 (36%), Positives = 28/46 (59%)
Frame = +1
Query: 615 EQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLP 660
E G A FNG N +C++ ET+ ++DE+++ I+ DE L+P
Sbjct: 589 ELGGASFNGEMNLNCNSLYETNDEVDEDEEEEID-----GDEDLIP 711
>TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC 6.4.1.2) -
potato chloroplast, partial (66%)
Length = 5011
Score = 32.3 bits (72), Expect = 1.2
Identities = 47/220 (21%), Positives = 82/220 (36%), Gaps = 12/220 (5%)
Frame = +3
Query: 162 EDGNSQQNVKNVSMESSPFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPG 221
+ GN ++ S +P + ++ D + D DY+ LQK + EP+
Sbjct: 2724 DSGNDDDKLQKAS---DILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTLEPEND 2894
Query: 222 STTSVAYGVQERDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYR 281
+ T D I++ H ++E E + E ++ Y
Sbjct: 2895 NDTD--------DYDKIQRAHY------------------ILEPENDSDTEPDD-EPDYE 2993
Query: 282 NYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKI 341
E D + DD+ K +QKA D IL PE S + + S T + K+
Sbjct: 2994 PDDEPDDEPDDDDK-----LQKASD-ILEPENPSDSEKVQNSSEIESTGIMENPSDSEKV 3155
Query: 342 E---ERNTTTFTESPKEVPKMESK---------QKSWSHL 369
+ E +T E+P + K+++ +K +SHL
Sbjct: 3156 QNSSEIESTGIMENPSDSEKVQNSSEIESTGIIRKDFSHL 3275
Score = 29.3 bits (64), Expect = 9.7
Identities = 31/146 (21%), Positives = 60/146 (40%), Gaps = 1/146 (0%)
Frame = +3
Query: 626 NSSCHNYNETDSDMDE-EKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEK 684
N S NY+++D+D D+ + + + +QK D L PE ++ + D R++ E
Sbjct: 2781 NYSAENYSDSDNDSDDNDPDDDYDTLQKGTD-TLEPENDNDTDDYDKIQRAHYILEPEND 2957
Query: 685 SEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPREL 744
S+ E ++ + P + ++K KA D + NP ++
Sbjct: 2958 SDTEPDDEPDYEPDDEPDDEPDDDDK---------------LQKASDILEPENPSDSEKV 3092
Query: 745 PSDANFEGEKVFLNRQTSEERKKSEE 770
+ + E + N SE+ + S E
Sbjct: 3093 QNSSEIESTGIMENPSDSEKVQNSSE 3170
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 32.3 bits (72), Expect = 1.2
Identities = 30/110 (27%), Positives = 48/110 (43%)
Frame = +3
Query: 187 PSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYK 246
P ++E KDK E+ QK+ ++ E + +E D + KK YK
Sbjct: 483 PEEISEIIDVKKDK------EIKQKAKSESESEES---------KESDSELRKKRRKSYK 617
Query: 247 HAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKN 296
+ S++ E E E ED + S R YSE+DSD + E+++
Sbjct: 618 KSRESDS----------ESESESEVEDRKRRKS-RKYSESDSDTNSEEED 734
>AL370046 GP|10177686|db emb|CAB62594.1~gene_id:MUL8.6~similar to unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 351
Score = 32.0 bits (71), Expect = 1.5
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +3
Query: 1191 LRQVVSKLTPARKRKVELLVEAFETVV 1217
+ + SKL ARK KV+ LV AFETV+
Sbjct: 12 IEETASKLVEARKSKVKALVGAFETVI 92
>TC77395 similar to GP|4874305|gb|AAD31367.1| expressed protein {Arabidopsis
thaliana}, partial (65%)
Length = 1649
Score = 32.0 bits (71), Expect = 1.5
Identities = 35/189 (18%), Positives = 73/189 (38%), Gaps = 22/189 (11%)
Frame = +2
Query: 131 LKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTV 190
LK +K + KD ++K + K +++ + + + +V + + APP
Sbjct: 386 LKKKKKSDKDKKNKGSSKSTKSESESQAFWAPAPLNATSWADVDDDDDDYYLTTAPPQFS 565
Query: 191 NECDTSTKDKHMV---TDYEVLQKSSTQEE-----PKPGSTTSVAYGVQER--------- 233
+ + K +H ++ ++L +EE P+P V++
Sbjct: 566 DPQPSEDKPEHFEDTESEEDILDDGDEEEEEQDHEPEPEHAVKTEPEVKKHAEVPVMPKE 745
Query: 234 -----DQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDS 288
+K KK L A+L++ G + +KE G+ +E ++ +
Sbjct: 746 AERQLSKKERKKKELEELEALLADFG-------VTQKESNDGQSQDESQGVPQDKKGVEG 904
Query: 289 DMDDEKKNV 297
D+D EKK +
Sbjct: 905 DVDGEKKEI 931
>TC84931 SP|P75780|YBIL_ECOLI Probable tonB-dependent receptor ybiL
precursor. {Escherichia coli}, partial (38%)
Length = 868
Score = 31.6 bits (70), Expect = 2.0
Identities = 33/148 (22%), Positives = 58/148 (38%), Gaps = 5/148 (3%)
Frame = +2
Query: 499 VTERARNDKNMDACKKNDESATVKST----ATKAVKFPVCDTGIMEEEVTAEGEYKVQEK 554
++++ RND +DA SA++ S T V + DT + V E +
Sbjct: 14 ISKQPRNDSGIDA------SASIGSAWFRRGTLDVNQVIGDTTAVRLNVMGEKTHDAGRD 175
Query: 555 SIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVG-KDKEG 613
+ E ++G + + V +G+ ++ Y+ H+ D +P +G
Sbjct: 176 KVKNE--RYGVAPS-VAFGLGTANRLYLNYLHVTQHNTP-------DGGIPTIGLPGYSA 325
Query: 614 REQGDAVFNGGNNSSCHNYNETDSDMDE 641
G A N HN+ TDSD D+
Sbjct: 326 PSAGTAALNHSGKVDTHNFYGTDSDYDD 409
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 31.6 bits (70), Expect = 2.0
Identities = 28/95 (29%), Positives = 41/95 (42%), Gaps = 9/95 (9%)
Frame = +3
Query: 626 NSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSR----SYGSDEL 681
+SS + + DS DE+KK V + + E+E SSDD K SDE
Sbjct: 258 DSSDSDDEDDDSSSDEDKKPVAAKKE-------VSESESDSSDDEDKMNVDKDGSDSDES 416
Query: 682 LEKSEGE-----REEMNATSFTETPKEAKKTENKP 711
E+SE E ++++ + K KK N P
Sbjct: 417 EEESEDEPSKTPQKKIKDVEMVDAGKSGKKAPNTP 521
>TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1635
Score = 31.6 bits (70), Expect = 2.0
Identities = 35/133 (26%), Positives = 59/133 (44%), Gaps = 14/133 (10%)
Frame = +2
Query: 1077 NSTIDRTGGIYSEGLNQKEQKMESG-------NGIV--EERKEESVSKEVNKPNQKLSRN 1127
N + DRT I ++ N+ ++ ES + IV E+ E+ S + K NQKL +
Sbjct: 1106 NKSNDRTSEIANDSQNKGQKSSESFVWPDAAVSAIVALEKASHEAFSTDFQKYNQKLRQL 1285
Query: 1128 WSNLKKVVLLRRFIKALE----KVRKFNPREPRY-LPLEPDSEDEKVQLRHQDMAERKGT 1182
NLK LL R + E K+ P E + L E ++ E + +H M + + +
Sbjct: 1286 DFNLKNNALLARRLLNGELKPSKILNMTPIELKEGLTAEEKTKKEPDEKQHMQMTDARCS 1465
Query: 1183 EEWMLDYALRQVV 1195
LR+++
Sbjct: 1466 RCTDSKVGLREII 1504
>TC86700 similar to GP|18377835|gb|AAL67104.1 At1g24120/F3I6_4 {Arabidopsis
thaliana}, partial (83%)
Length = 1929
Score = 30.8 bits (68), Expect = 3.3
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +1
Query: 119 PLKRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTK 157
PLKR S + R S K +K+DG +++ N R+ +K K
Sbjct: 1279 PLKRSNSSKSRSKTSLKESKEDGETREKRNTRERPRKKK 1395
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,068,337
Number of Sequences: 36976
Number of extensions: 373979
Number of successful extensions: 2009
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 1849
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2000
length of query: 1222
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1115
effective length of database: 5,058,295
effective search space: 5639998925
effective search space used: 5639998925
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC125478.8


